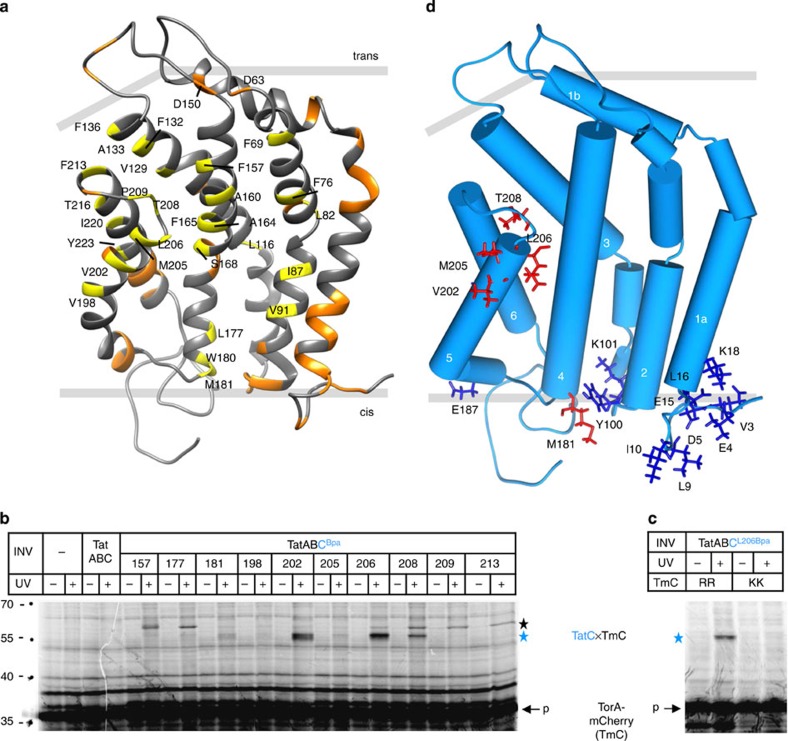
Figure 1. Contacts between TatC and a membrane-inserted RR-precursor.
(a) Model of E. coli TatC based on the structure of A. aeolicus TatC7 using PDB code 4B4A. Indicated in yellow are all amino acids replaced by Bpa in this study, and in orange those of a previous analysis19. (b) The model RR-precursor TorA-mCherry was synthesized and radioactively labelled by in vitro transcription/translation in the absence or presence of inverted E. coli inner membrane vesicles (INV). In addition to TatA and TatB, INV contained either wild-type TatC (TatABC) or the indicated Bpa variants of TatC (TatABCBpa). In samples labelled (+), crosslinking was initiated by irradiation with ultraviolet light. Radiolabelled translation products were separated by SDS–PAGE and visualized by phosphorimaging. Indicated are the positions of molecular size standard proteins (left-hand side), the TorA-mCherry (TmC) precursor (p), and the crosslinked TatC–TmC complex (blue star). The black star marks a ultraviolet light-dependent crosslink of unknown nature between TorA-mCherry and several TatC variants. (c) comparing crosslinking of the L206Bpa variant of TatC to wild-type TorA-mCherry (RR) and to a mutant with an inactive signal peptide (KK). (d) Highlighted in red are all residues that in b showed distinct contacts to TorA-mCherry when replaced by Bpa, and in blue the precursor contact sites identified in our previous study19. The helices of TatC are numbered.