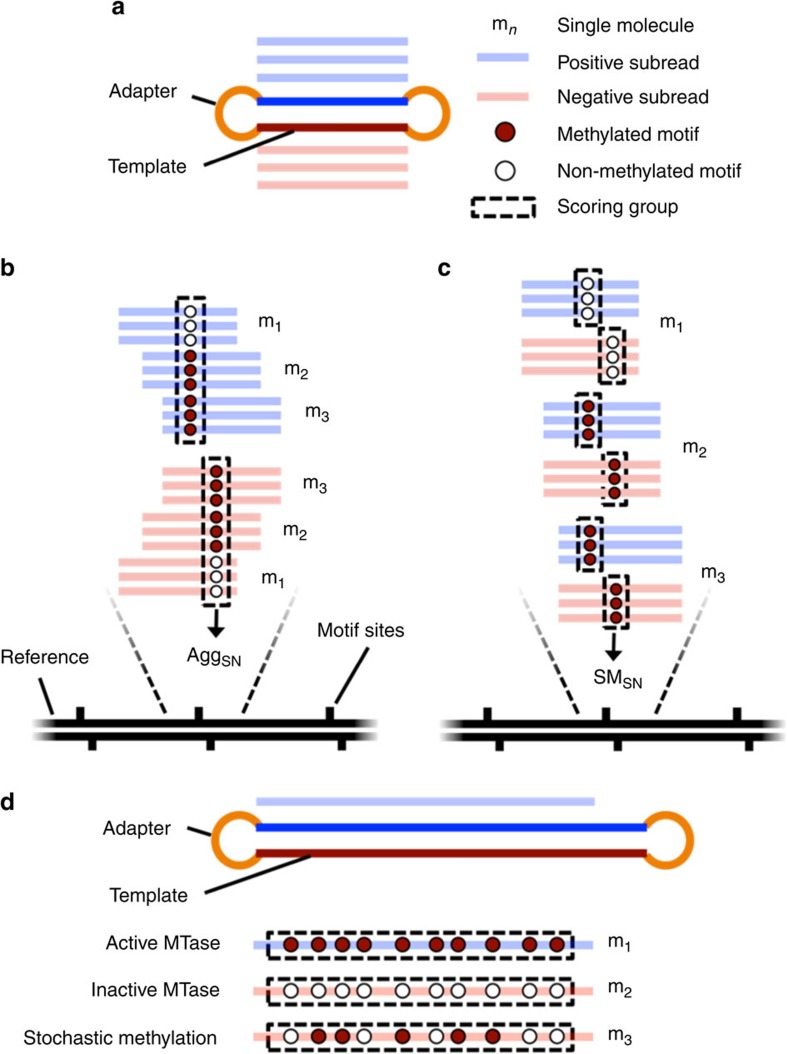
Figure 1. SMALR methods for methylation detection in SMRT reads.
Schematic illustrating the general approaches of both the existing and two proposed SMALR methods for detecting DNA methylation in SMRT sequencing reads. (a) A single SMRT sequencing molecule (short DNA insert+adapters) and the subreads that are produced during sequencing. (b) The existing methylation detection method is based on a molecule-aggregated, single-nucleotide (AggSN) score. For a given strand and genomic position, the IPD values from all the subreads aligning to that strand and position are aggregated together across all molecules to infer the presence of a consensus methylated base. (c) The proposed single molecule, single nucleotide (SMSN) method for detecting DNA methylation relies instead on separate consideration of subreads from different molecules. The SMSN scores are calculated for each molecule, strand and genomic position. (d) A single SMRT sequencing molecule (long DNA insert+adapters) with a single long subread and the proposed single molecule, pooled (SMP) approach for assessing MTase activity. This approach pools together IPD values from multiple motif sites along the length of a single long subread.