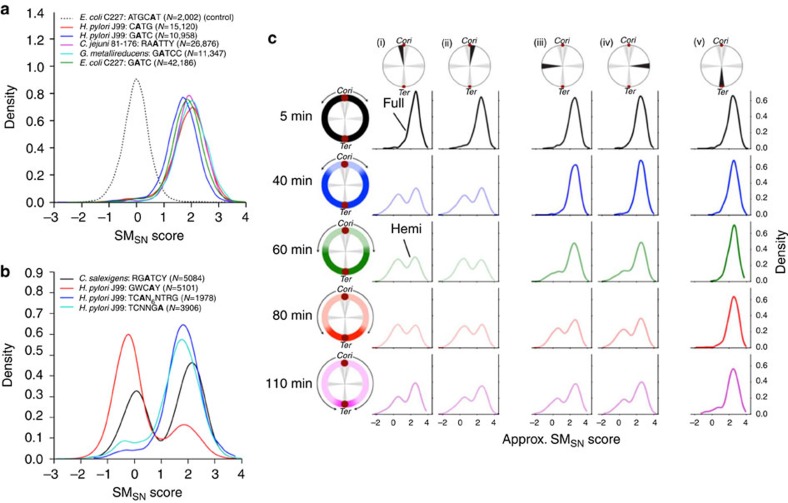
Figure 3. SMSN score distributions reveal epigenetic heterogeneity.
(a) Single molecule, single nucleotide (SMSN) score distributions for multiple bacterium-motif pairs (and the genome-wide motif count, N, of each motif) that exhibit near complete methylation, along with a non-methylated motif for comparison. (b) SMSN score distributions for multiple bacterium-motif pairs that display significant non-methylated fractions. The H. pylori J99 motifs show minor variation in the SMSN associated with each peak due to subtle differences in the chemistry version used for SMRT sequencing of the native and WGA samples. (c) SMSN interrogation of 5′-GANTC methylation at five genomic positions (columns) in a synchronized C. crescentus culture during a single round of DNA replication. Five time points (minutes post-synchronization; rows) provide snapshots of the bidirectional progression of the replication forks from the origin of replication (Cori) to the terminus (Ter). Grey wedges in the chromosome schematics show the 200-kb genomic regions where the SMSN scores are queried for each time point. Two regions are on either side of the Cori: (i) Cori - 0.1 Mbp and (ii) Cori+0.1 Mbp. Another two are halfway between Cori and Ter: (iii) Cori - 1 Mbp and (iv) Cori+1 Mbp. The final region covers the terminus: (v) Ter. Light (hemimethylated) to dark (fully methylated) colour shading in the schematic illustrates the approximate position of the replication fork at each time point. The bimodal distributions of approximate SMSN scores (Methods) reveal the progressive hemi-methylation of 5′-GANTC sites following the passage of the replication forks. Hemimethylated sites cannot transition back to full methylation until the MTase gene, ccrM, is transcribed, which does not occur until late in the replication process.