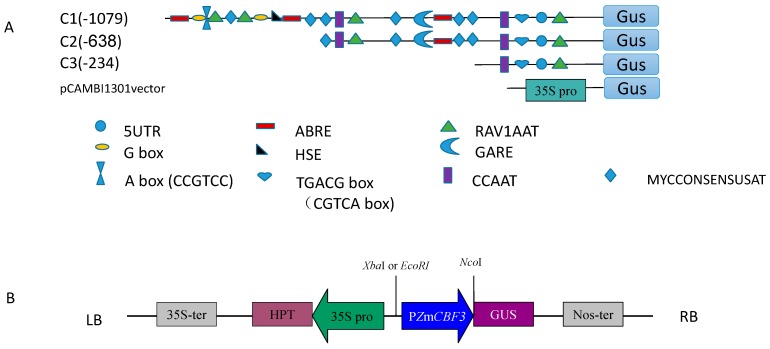
Figure 2.
Transformation of Arabidopsis plants using PZmCBF3:GUS constructs. (A) Schematic representation of the PZmCBF3 promoter constructs used to analyze GUS expression in Arabidopsis leaves. The serial 5′-deletion constructs of the PZmCBF3 promoter were fused to the GUS reporter gene in the vector, pCAMBIA1301; (B) Schematic representation of the PZmCBF3:GUS construct. The insertion position of the ZmCBF3 promoter in the vector is indicated with restriction enzyme sites (XbaI or EcoRI and NcoI). LB, left border; RB, right border; 35S-ter, CAMV 35S terminator; 35S pro, CAMV 35S promoter; GUS, β-glucuronidase gene; HPTII, hygromycinphosphotransferase (II) coding region; NOST, nopaline synthase terminator; PZmCBF3, ZmCBF3 promoter.