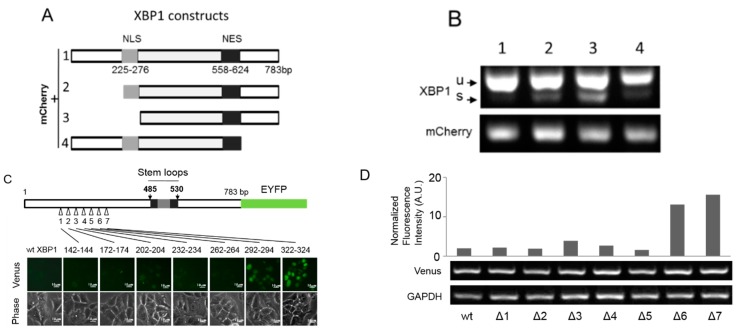
Figure 3.
Effects of flank sequences of stem-loop pair on the splicing of XBP1 mRNA. (A) Constructs of mCherry followed by XBP1 fragments. The number indicates the position of nucleotides in unspliced XBP1. NLS, putative nuclear localization sequence; NES, putative nuclear export sequence; (B) The total RNA of MCF-7 cells expressing the constructs in (A) was subjected to RT-PCT with XBP1 primers and mCherry primers; (C) Effect of 5′-nucleotide sequence of XBP1 mRNA on its basal splicing (scale bar = 10µm). XBP1-EYFP was constructed as ERAI1-783, as described in Figure 1A. ERAI1-783 cDNAs with Different deletions in the 5′-end sequence of XBP1 were respectively stably expressed in MCF-7 cells. Δ1, 2, 3, 4, 5, 6, and 7 represent seven deletion mutants. Fluorescence images indicate the basal unconventional splicing; and (D) The normalized fluorescence intensity in (C) was presented as the ratio of fluorescence intensity/ERAI1-783 expression level. Venus RT-PCR indicates the expression level of ERAI1-783 constructs. The value is obtained from the mean of one representative triplicate experiment.