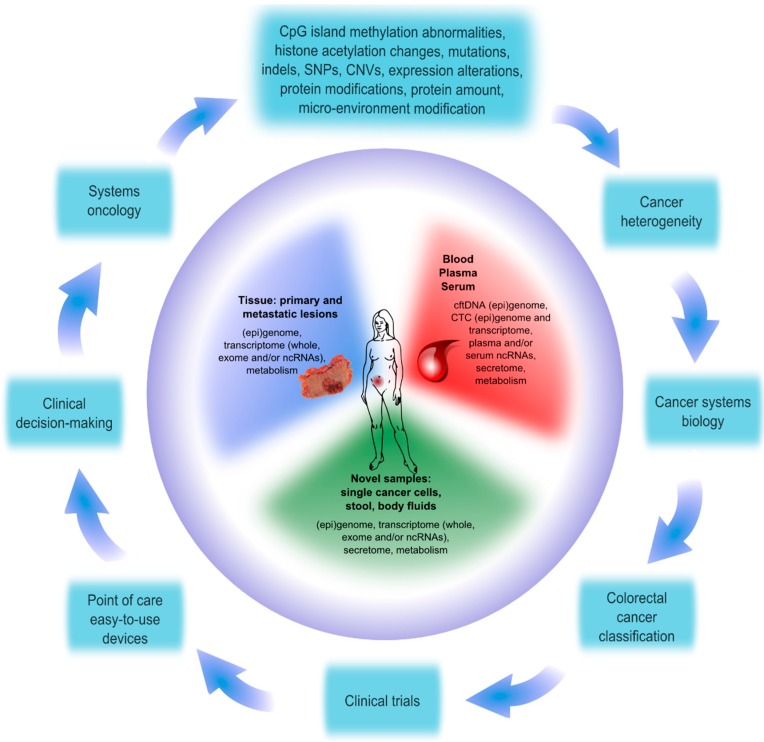
Figure 1.
The study of colorectal cancer can be addressed using many different sample sources beyond biopsies from primary tumors. Additional sample sources include metastatic tissue, blood, and derivatives (plasma and serum), as well as other body fluids, stool or single cancer cells. Using high throughput technologies, within these samples we can analyze tumor DNA, circulating free DNA (cftDNA), circulating tumor cells (CTCs), RNA and proteins in order to discover alterations in (epi) genome, transcriptome, secretome, metabolism, and so on. These alterations include CpG island methylation, histone modification, mutations, insertion and deletion events (indels), single nucleotide polymorphisms (SNPs), copy number variations (CNVs), changes in the amount of proteins and RNAs, and so on. All these alterations contribute together to tumor heterogeneity, and to improve the overall understanding of colorectal cancer, cancer systems biology should integrate and link them to each other. Once integrated, we will be able to develop molecular classifications to better define the possible outcome scenarios and the therapeutic strategies to follow with each patient individually. Each classification should be tested in properly designed clinical trials, and in the future, if the classification demonstrates usefulness, point-of-care devices could be developed in order to apply the novel tools to facilitate clinical decision-making. This would be a definite step towards the implementation of systems oncology, which should be continually improved with more analyses in more samples.