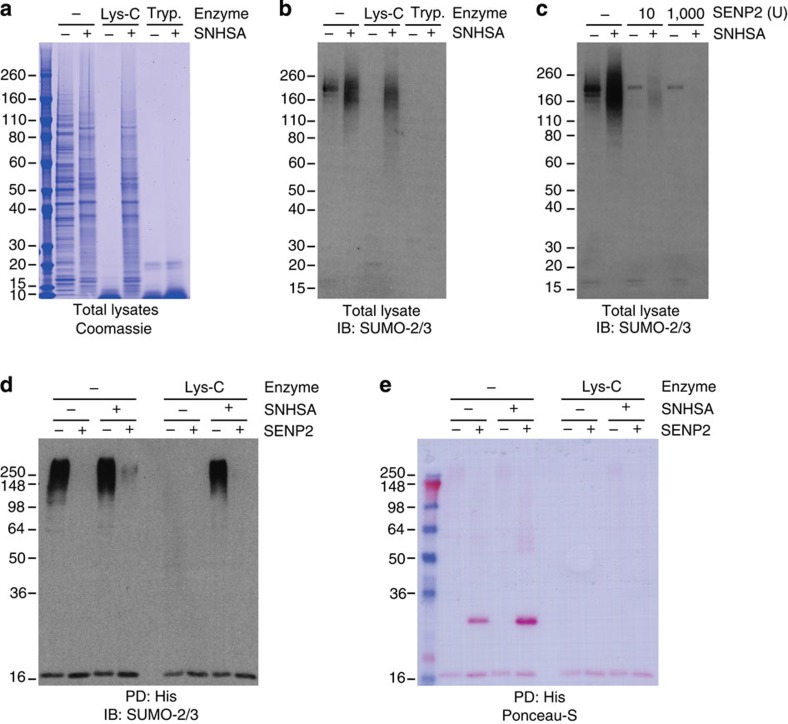
Figure 3. SENP2 is able to remove acetylated SUMO from acetylated proteins after SNHSA treatment.
(a) HeLa cells were lysed in 8 M urea, homogenized and subsequently treated with 10 mM SNHSA or mock treated. Next, both lysine-blocked and control samples were treated with either trypsin or Lys-C, or mock treated. All samples were size-separated by SDS–PAGE, and total protein content was visualized using Coomassie. The experiment was performed in biological duplicate. (b) As in a, but with samples transferred to membranes after SDS–PAGE, subsequently probed using a SUMO-2/3 antibody and visualized using chemiluminescence. (c) HeLa cells were lysed in 8 M urea, homogenized and subsequently treated with 10 mM SNHSA or mock treated. Next, both lysine-blocked and control samples were treated with either a standard or large amount of SENP2, or mock treated. All samples were size-separated by SDS–PAGE, transferred to membranes, probed using a SUMO-2/3 antibody and visualized using chemiluminescence. The experiment was performed in biological duplicate. (d) HeLa cells stably expressing lysine-deficient His10-tagged SUMO-2 were lysed and homogenized and subsequently SUMOylated proteins were enriched by Ni-NTA pulldown (PD:His). SUMOylated proteins were treated on-beads with SNHSA or mock treated, prior to elution. After elution, proteins were either treated with SENP2 or mock treated. Finally, samples were digested using Lys-C or mock digested. All samples were size-separated using SDS–PAGE, transferred to membranes, probed using a SUMO-2/3 antibody and visualized using chemiluminescence. The experiment was performed in biological duplicate. (e) As in d, but with total protein content visualized using Ponceau-S staining.