Figure 3.
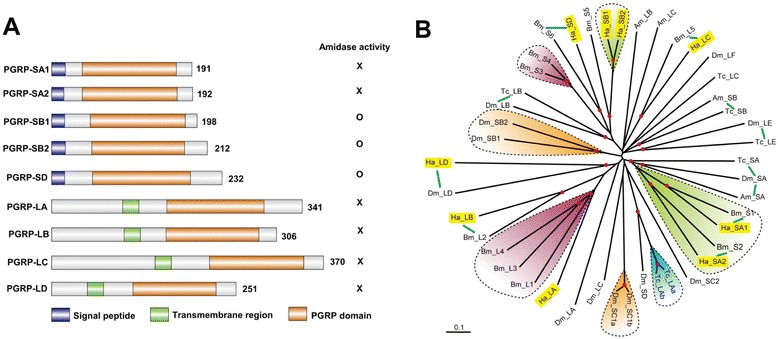
The PG recognition proteins of H. armigera. (A) Schematic representations of the H. armigera PGRP domain structures. Lengths of the amino acid sequences are indicated. (B) Phylogenetic analysis of PGRPs. The amino acid sequences of 9 H. armigera (Ha), 7 T. castaneum (Tc), 13 D. melanogastor (Dm), 11 B. mori (Bm), and 4 A. mellifera (Am) PGRPs are compared. Scale bar, 0.1 substitutions per site. The tree shows that HaPGRP and BmPGRPs form good orthologous groups, except for HaPGRP-LD. Red dots at nodes indicate bootstrap values greater than 800 from 1,000 trials. The putative 1:1 or 1:1:1 orthologs were connected by green lines. HaPGRP-LA, −LB, −LC, and -LD contain a transmembrane domain, while HaPGRP-SA1, −SA2, −SB1, −SB2, and -SD have a signal peptide. HaPGRP-SB1,-SB2, and -SD contain the key residues of the amidase activity that hydrolyzes PGs.
