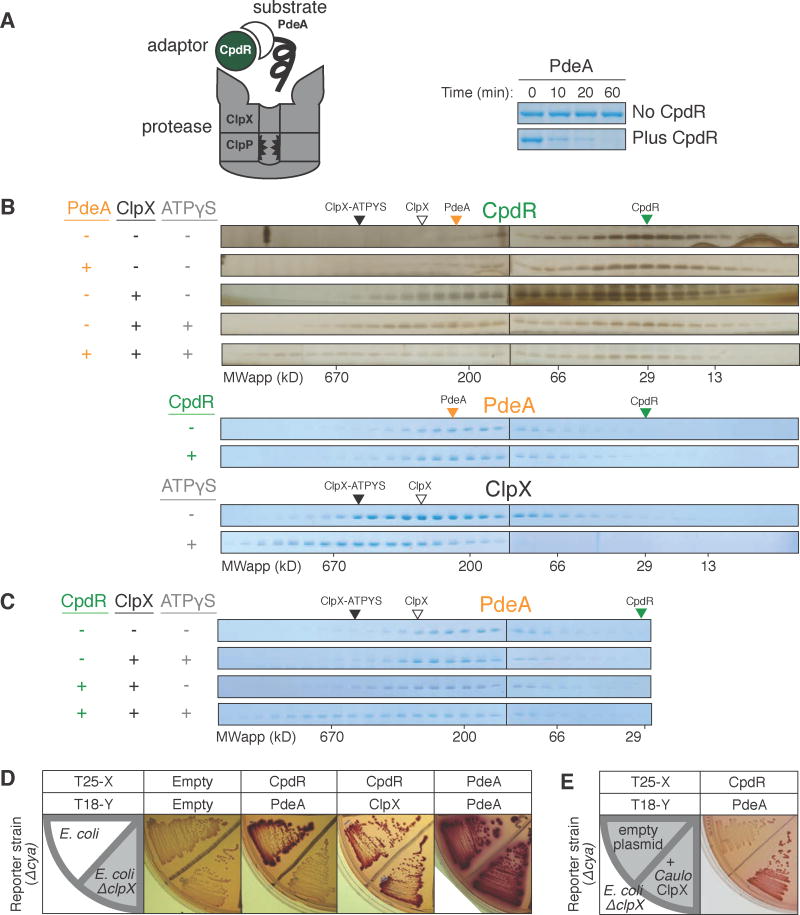
Figure 1. Interaction between adaptor CpdR and unfoldase ClpX enables recruitment of substrate PdeA.
(A) Degradation of PdeA by ClpXP when mediated by CpdR in the presence of an ATP regeneration system and 1mM GTP (which enhances PdeA degradation (Abel et al., 2011)). (B) Size exclusion chromatography (SEC) profiles of CpdR (±PdeA/ClpX/ATPγS). Profiles for PdeA (±CpdR) and ClpX (±ATPγS). Robust detection of CpdR required silver staining as it stained poorly by Coomassie (Figure S1). (C) SEC profiles of PdeA (±CpdR/ClpX/ATPγS). Colored triangles mark peak locations of individual proteins. (D) BACTH assay using McConkey agar results in red colonies when interacting proteins are fused to complementary fragments of adenylate cyclase (cya), T18 and T25 (Karimova et al., 1998). Fusions of CpdR, PdeA and ClpX (C. crescentus ortholog) examined in E. coli wild-type and ΔclpX (shaded) reporter strains. (E) Interaction between CpdR and PdeA examined in the ΔclpX reporter strain harboring a plasmid expressing C. crescentus ClpX (pCL1920-ClpX) or empty plasmid (pCL1920). For (B&C), 20μM CpdR/5μM PdeA/5μM ClpX6/5mM(sample mixture) or 1mM (in running buffer) ATPγS were used, with 1mM GTP throughout. See also Figure S1.