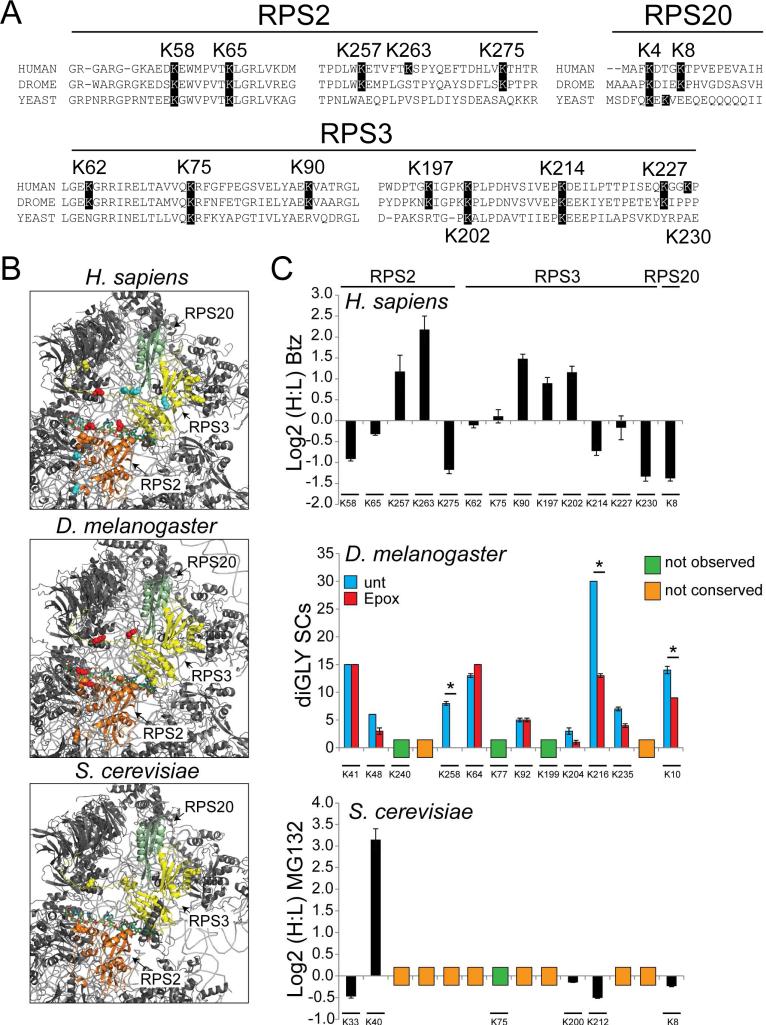
Figure 7. 40S regulatory ubiquitylation is conserved across eukaryotes.
A) Multiple sequence alignments of segments of RPS2, RPS3, and RPS20 from H. sapiens, D. melanogaster, and S. cerevisiae that contain ub-modified lysine residues. The highlighted lysine positions indicate lysine residues observed to be ub-modified in human cells (position is human numbering).
B) Structural representation of the solvent exposed surface of the 40S subunit from H. sapiens (PDB:4V6X), D. melanogaster (PDB:1V6W), and S. cerevisiae (PDB:4V7R). Representations and coloring are as in Fig 2C.
C) Top: SILAC (H:L) Log2 ratios for selected RPS2, RPS3, and RPS20 ub-modified peptides from HCT116 cells treated with bortezomib for 8 hours. Primary data is extracted from published datasets (Kim et al., 2011). Error bars represent SEM from multiple peptide MS quantifications. Middle: Spectral counts (SCs) for ub-modified peptides from untreated (blue bars) and epoxomicin treated (red bars) Drosophila S2 cells. Error bars represent SEM from triplicate measurements. * indicates a p-value of < 0.05 using Student's t-test. Bottom: SILAC (H:L) Log2 ratios for selected diGly-modified peptides from heavy labeled yeast cultures treated with MG132 prior to mixing with untreated, unlabeled control cells. Error bars represent SEM from multiple peptide MS quantifications. The position of the ub-modified lysine is aligned to the analogous human position. The lysine position within the D. melanogaster or S. cerevisiae 40S protein sequences is shown below. Lysine residues not found to be ub-modified or conserved are indicated.