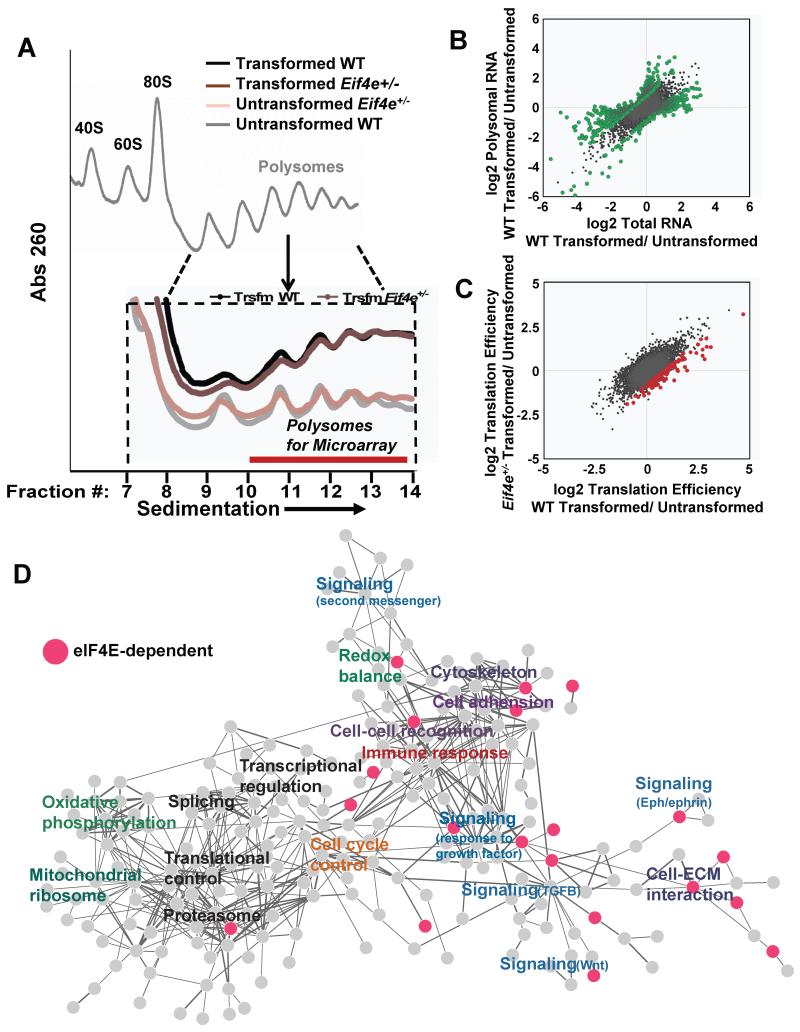
Figure 3. eIF4E is required for expression of the oncogenic translational program.
A) Representative polysome profiles of transformed and untransformed WT and Eif4e+/− MEFs separated on a sucrose gradient. Inset highlights the polysomal fractions (10-13) used for translational profiling by polysomal microarray. B) Changes in total RNA and polysome associated RNA in WT MEFs upon oncogenic transformation by HRasV12 and Myc. Green points indicate genes with a fold change in translation efficiency (polysomal/total RNA) greater than 1.7 fold, p<0.05 (See Table S3). C) Changes in translation efficiency (TE) induced by transformation in WT and Eif4e+/− MEFs. Red points indicate genes whose fold change in TE during transformation is reduced in Eif4e+/− MEFs compared to WT MEFs (>1.7 fold decrease in TE, p<0.05) (See Table S2). D) Network-based cluster analysis of genes and associated functional classes whose translation is increased upon transformation in WT MEFs. Pink nodes represent genes whose translation is impaired in transformed Eif4e+/− cells. See also Figure S3,