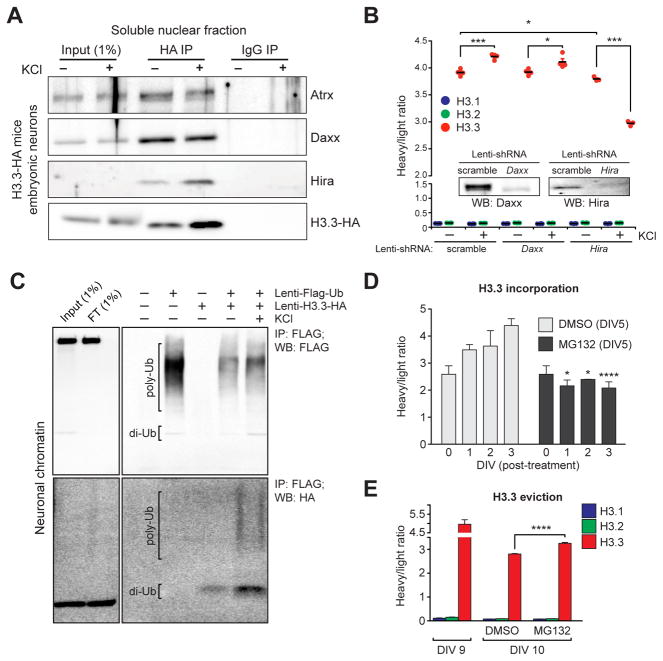
Figure 4. Neuronal histone dynamics are Hira-dependent and require active proteasomal degradation.
(A) Immunoprecipitations of free H3.3-HA from embryonic neuronal soluble nuclear fractions following depolarization. IgG controls indicate selective immunoprecipitations of H3.3 interacting proteins. (B) LC-MS/MS SILAC analysis of H3.1/2 vs. H3.3 chromatin incorporation −/+ KCl stimulation (5 hrs) in mouse embryonic neurons transduced with one of three lentiviruses (scramble, shRNA-Daxx or shRNA-Hira) (n = 3–4 biological replicates/group). Insert: western blot analysis indicating knockdown of targeted proteins vs. a scrambled control. (C) H3.3 polyubiquitylation in response to KCl depolarization in embryonic neuronal chromatin. Primary neurons were transduced with lenti-Flag-Ub, lenti-H3.3-HA or both, and Flag-tagged ubiquitin was immunoprecipitated from fractionated neuronal chromatin followed by Western blot analysis for ubiquitin (Flag) and ubiquitylated H3.3 (HA). (D) LC-MS/MS SILAC analysis of H3.3 chromatin incorporation (beginning at DIV 5 of SILAC labeling) in mouse embryonic neurons following treatment with DMSO (control) or MG132 (proteasome inhibitor). Cells were analyzed at DIV 0, 1, 2 and 3 post-treatment (n = 3 biological replicates/group). (E) LC-MS/MS SILAC analysis of H3.3 chromatin eviction (beginning at DIV 9 of SILAC labeling) in mouse embryonic neurons following treatment with DMSO or MG132 (n = 3–5 biological replicates/group). Data represented as means ± SEM. *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001. See also Figure S4.