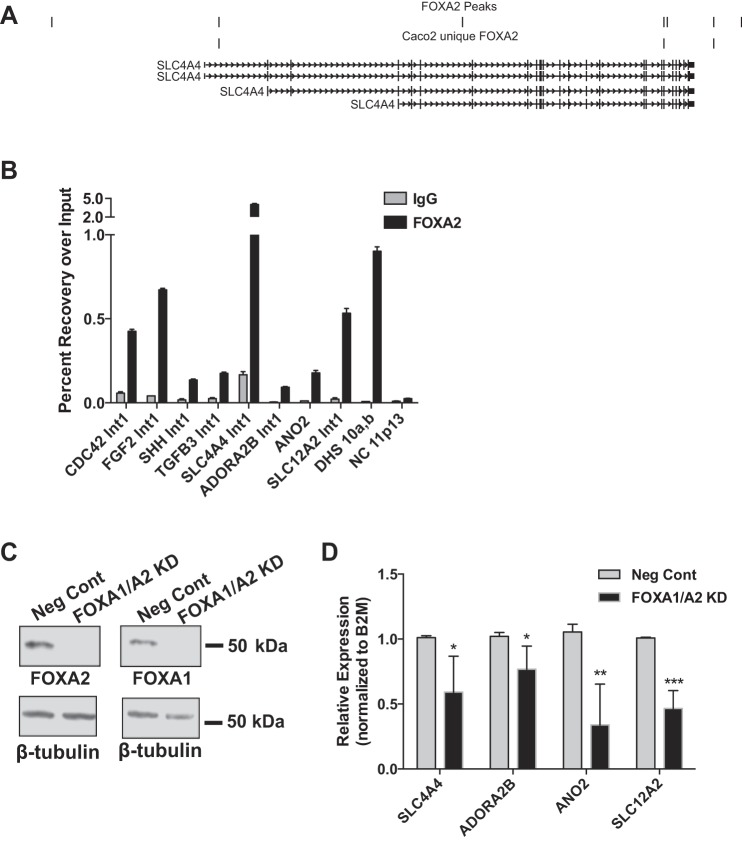
Fig. 2.
FOXA2 regulates gene targets identified by ChIP-seq. A: UCSC genome browser view of the SLC4A4 locus showing all FOXA2 peaks and those unique to Caco2 datasets. B: representative ChIP-qPCR validation of FOXA2 binding sites at targets enriched in the GO terms cell morphogenesis, tube development, and plasma membrane part. NC 11p13 is a negative control region with no FOXA motifs and no known regulatory function. C: representative Western blot showing Caco2 cells treated with either a negative control (Neg Cont) siRNA or siRNAs targeting both FOXA1/A2 [FOXA1/A2 knockdown (KD)]. β-Tubulin is shown below as a loading control. D: RT-quantitative (q)PCR analysis of selected target genes after targeting with NC or FOXA1/A2 siRNAs. All transcripts are normalized to β-2-microglobulin (B2M) levels and the data shown are from >3 biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001.