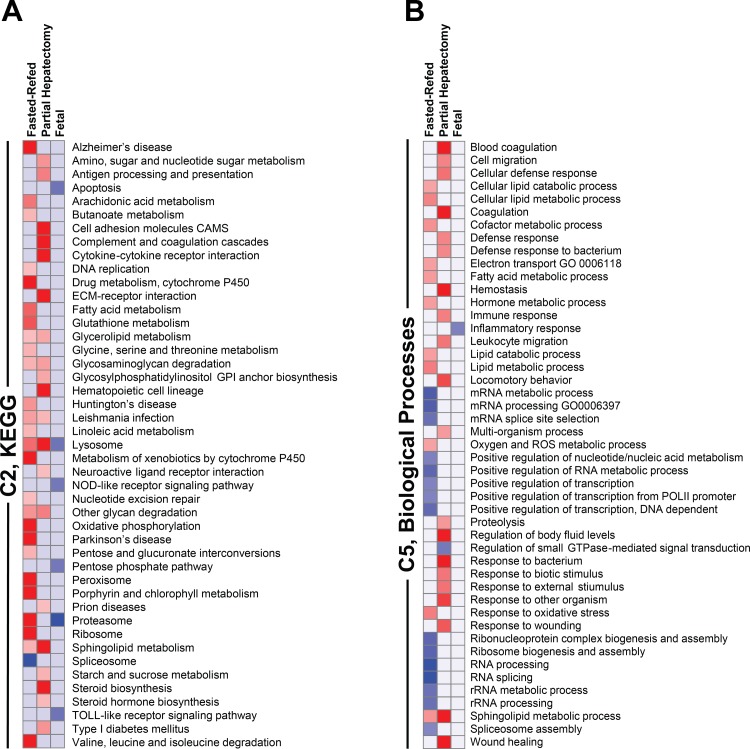
Fig. 4.
Gene set enrichment analysis (GSEA) of rapamycin effect on the transcriptome in adult and fetal liver. Results are depicted as heat maps for the 3 experiments: fasting-refeeding in adult rats, partial hepatectomy in adult rats, and late-gestation fetal rats. The heat maps depict the response to rapamycin. All gene sets that were significant at Q < 0.01 are shown. Those that were enriched for upregulated genes are shown in red, and those that were enriched for downregulated genes are in blue. The intensity of the color is proportional to the degree of significance based on false discovery rate (FDR) Q value. All gene sets that were significant for any experiment are shown. Data are shown for the C2 Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets (A) and the C5 biological processes gene sets (B). ECM, extracellular matrix; NOD, nucleotide-binding oligomerization domain; GO, gene ontology; ROS, reactive oxygen species.