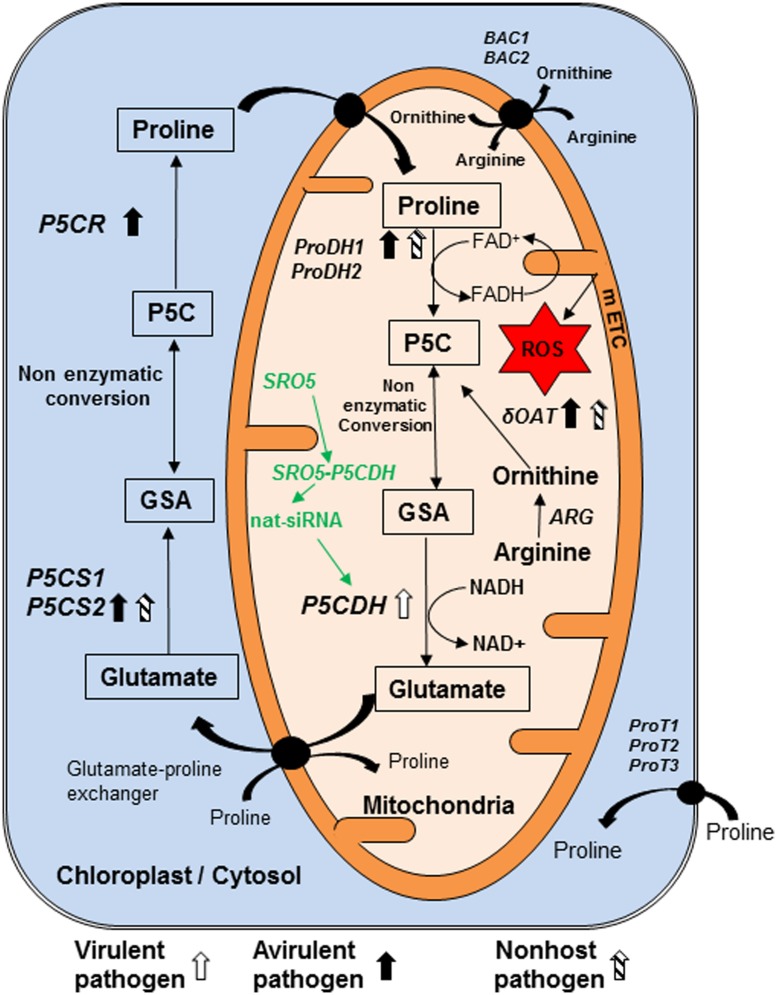
FIGURE 1.
Model showing genes and pathways possibly involved in synthesis and catabolism of P5C in plant cell and their regulation in response to pathogen infection. Pyrroline 5-carboxylate (P5C) is the intermediate product of both biosynthesis and catabolism of proline. It is synthesized in mitochondria during catabolism of proline by enzyme proline dehydrogenase (ProDH1/2). We speculate that like their counterparts from bacteria and yeast, this enzyme reduces FAD+ to FADH and increases electron flow in mitochondrial electron transport chain (mETC). Arginine is converted into ornithine by arginase (ARG) enzyme. Another enzyme delta-ornithine amino transferase (δOAT) convert ornithine to P5C in mitochondria. P5C is catabolized by pyrroline 5-carboxylate dehydrogenase (P5CDH) in mitochondria into glutamate. In addition, P5C is synthesized in cytosol and chloroplast, from glutamate by pyrroline 5-carboxylate synthase 1 and 2 (P5CS1, P5CS2) and converted to proline by pyrroline 5-carboxylate reductase (P5CR). P5C and glutamate semi aldehyde (GSA) are non-enzymatically inter-convertible forms. Virulent pathogen infection in plants increases transcript accumulation of ProDH1. Avirulent pathogen infection increases transcript accumulation of P5CS2 and ProDH1. Non-host pathogen infection increases transcript accumulation of δOAT as well as ProDH1 and P5CS2. Three transporters located on plasma membrane are ProT1, ProT2 and ProT3. Arginine is imported into mitochondria directly and/or in exchange of ornithine by basic amino acid career (BAC1) and BAC2. P5CDH gene is down regulated post transcriptionally by natural siRNAs from similar to RCD one-5 (SRO5) genes. No direct evidence available for this pathway shown in green color during plant–pathogen interaction and this information is speculated based on evidence available under salt stress. Compounds shown in rectangle are important component of metabolism of P5C and line arrows indicates the direction of synthesis. Curved arrows show the transport of compounds. Transporters shown in circle are present on membrane. Block thick arrows show upregulation of genes, in which white arrow indicates virulent pathogen, dark arrows represent avirulent pathogen and striped arrows indicate non-host pathogen. ROS, reactive oxygen species. This model is predominantly based on information from Arabidopsis thaliana literature (Ayliffe et al., 2002; Borsani et al., 2005; Miller et al., 2009; Verslues and Sharma, 2010; Cecchini et al., 2011; Senthil-Kumar and Mysore, 2012).