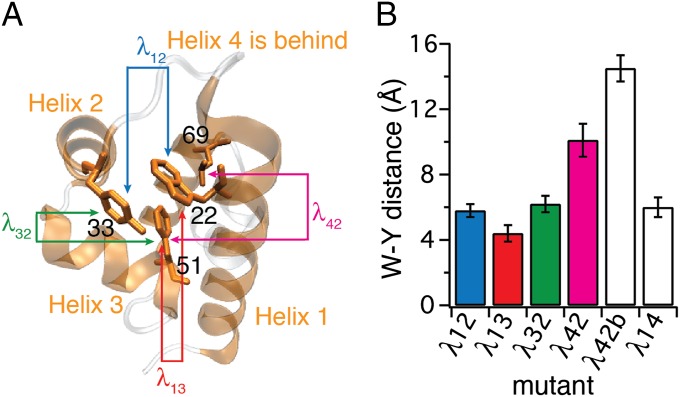
Fig. 1.
Construction of λ6–85 mutants. (A) Crystal structure of the template mutant λ12 (PDB ID code 3KZ3) with residues 22, 33, 51, and 69 highlighted in helices 1, 2, 3, and 4, respectively. Helices 1 through 4 in which mutations were made are shown in orange; the rest of the sequence is shown in gray. Arrows between pairs of residues and mutant names λij follow the coloring and naming scheme used throughout the paper. Mutants λij are named according to the helix pair contact probed. (B) Tryptophan–tyrosine (W-Y) distances calculated for six λ6–85 mutants. Error bars represent the SD from fluctuations of the protein structures in the 100 ns of native-state MD simulations (SI Appendix, Fig. S1 and Table S2). All sequences are in SI Appendix, Fig. S1.