Fig. 1.
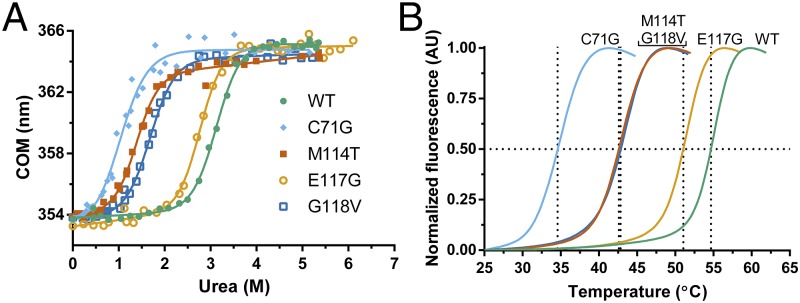
ALS-linked mutations destabilize PFN1. Chemical and thermal denaturation studies reveal that ALS-linked variants C71G, M114T, and G118V, but not E117G, are severely destabilized relative to PFN1 WT. (A) Equilibrium unfolding curves for PFN1 WT and ALS-linked variants generated by measuring the intrinsic tryptophan fluorescence of the indicated protein equilibrated in increasing concentrations of urea. Data were processed to obtain the center of mass (COM) of the emission spectrum and then fit to a two-state model for protein folding. The resulting fits are displayed as solid lines. The corresponding thermodynamic parameters obtained from the fitted data are shown in Table 1. (B) Thermal denaturation profiles of PFN1 proteins measured by SYPRO Orange fluorescence as a function of increasing temperature were used to determine the apparent Tm, which is the temperature corresponding to 0.50 fluorescence signal as denoted by the intersection of the dashed lines for each curve.
