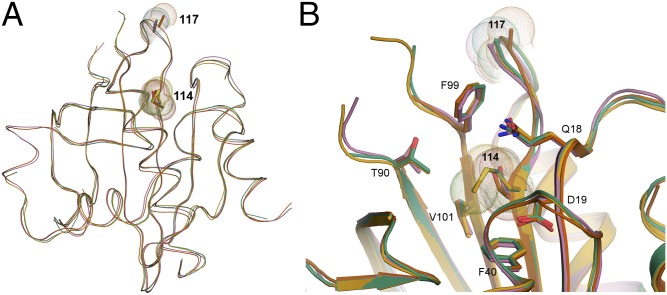
Fig. 3.
Superimposition of the crystal structures for PFN1 WT, E117G, and M114T. (A and B) The secondary and tertiary structures for PFN1 WT (green), E117G (mustard), M114T chain A (pink), and B (red) are highly superimposable. For each structure, sticks and spheres denote the side chains and van der Waals radii, respectively, for residues at position 114 and 117. Residue 117 is located within a solvent-exposed flexible loop that has no discernible secondary structure, whereas Met114 is located within a β-sheet toward the interior of the protein. (B) A zoomed cartoon representation showing residues within 4 Å of residue 114. The side chains of these residues are indicated as sticks with nitrogen, oxygen, and sulfur atoms indicated in blue, red, and yellow, respectively. The van der Waals radii of the atoms comprising residue 114 are reduced upon mutation of methionine (green and mustard structures) to threonine (red and pink structures).