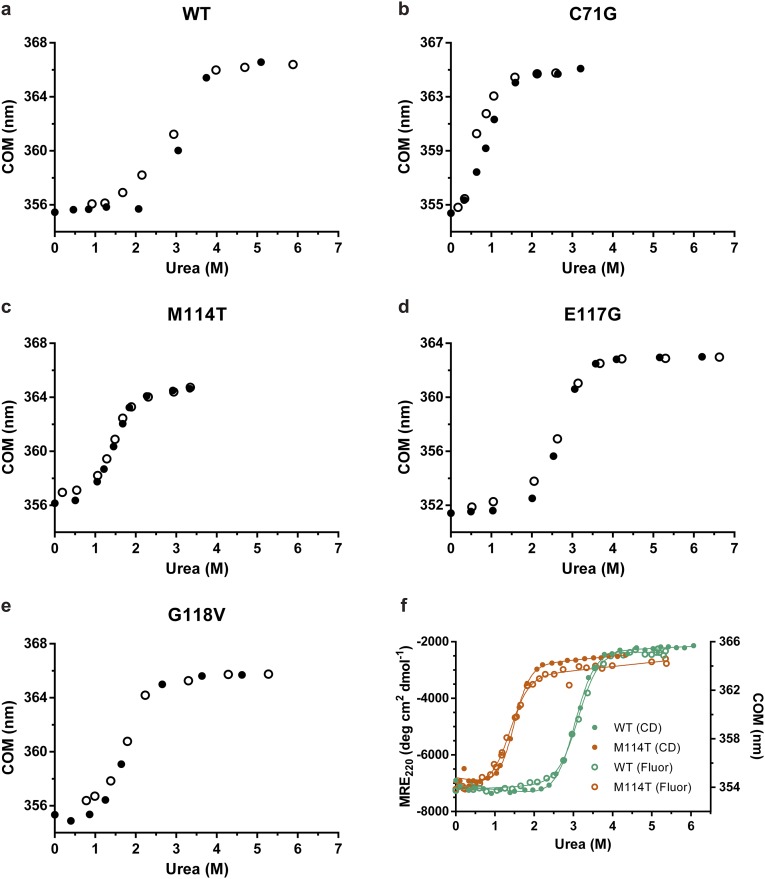
Fig. S2.
All PFN1 variants unfold by a two-state process. (A–E) PFN1 variants denatured in urea were refolded by diluting the urea. The final concentration of PFN1 in each sample was 10 μM and tryptophan fluorescence was used to monitor folding. The equilibrium transition regions overlay closely for the unfolding and refolding curves, indicating that the unfolding reaction is reversible. Filled and open circles represent unfolding and refolding, respectively. (F) The two-state unfolding of PFN1 observed by intrinsic fluorescence (data from Fig. 1A; Fluor) was verified by CD measurements for PFN1 WT and M114T. The concentration of protein used was 2 μM and 10 μM for tryptophan fluorescence and CD measurements, respectively. The y axis on the left is the mean residue ellipticity at 220 nm (MRE220) obtained from CD experiments, whereas the y axis on the right reflects the change in the COM (as shown in Fig. 1). The thermodynamic parameters obtained by fitting the CD data agree well with those obtained from the fluorescence data (Table 1) and are as follows: for WT ΔG° = 7.16 ± 0.11 kcal⋅mol−1, m = 2.36 ± 0.04 kcal⋅mol−1⋅M−1, Cm = 3.03 ± 0.07 M; for M114T ΔG° = 4.35 ± 0.10 kcal⋅mol−1, m = 2.95 ± 0.06 kcal⋅mol−1⋅M−1, Cm = 1.47 ± 0.05 M.