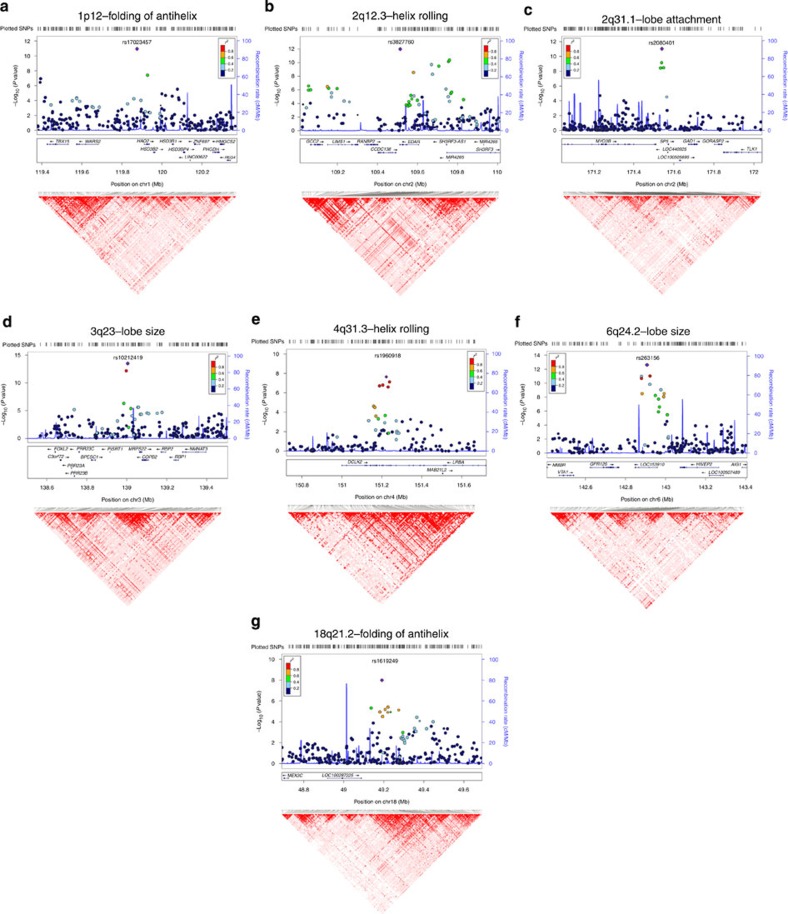
Figure 3. LocusZoom and linkage disequilibrium plots of significantly associated genetic regions.
Plots of the seven genomic regions showing genome-wide significant association to pinna traits (Table 1). For regions showing association with several pinna traits, we present here only results for the trait with strongest association (plots for the other associated traits are presented in Supplementary Fig. 7). Association results from a multivariate linear regression model (on a −log10 P scale; left y axis) are shown for SNPs ∼500 kb on either side of the index SNP (that is, the SNP with the smallest P value, purple diamond; Table 1) with the marker (dot) colour indicating the strength of LD (r2) between the index SNP and that SNP in the 1000 Genomes AMR data set. Local recombination rate in the AMR data is shown as a continuous blue line (scale on the right y axis). Genes in each region, their intron–exon structure, direction of transcription and genomic coordinates (in Mb, using the NCBI human genome sequence, Build 37, as reference) are shown at the bottom. Plots were produced with LocusZoom56. Below each region we also show an LD heatmap (using D′, ranging from red indicating D′=1 to white indicating D′=0) produced using Haploview57. Note that the location of SNPs on the LD heatmap can be shifted relative to the regional display on top of it.