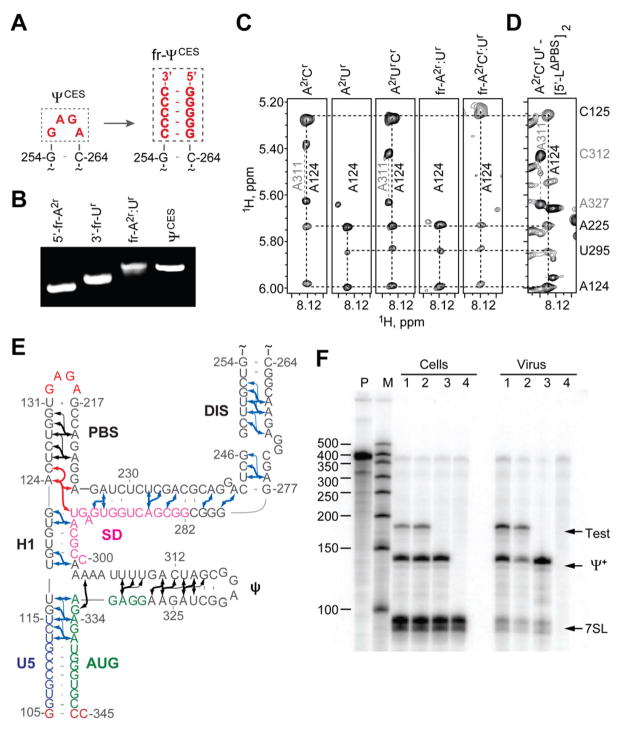
Figure 2.
Fragmentation-based 2H-edited NMR approach and observed ΨCES secondary structure. (A) The DIS loop of ΨCES served as the fragmentation site and was substituted by a stretch of intermolecular G-C base pairs. (B) Fragment annealing efficiency as measured by native polyacrylamide gel electrophoresis. (C) 2D NOESY spectra of uniformly labeled A2rCr-, A2rUr-, and A2rCrUr-ΨCES and segmentally-labeled fr-A2r:Ur- and fr-A2rCr:Ur-ΨCES samples used to make long-range NOE assignments. (D) Similarities in NOESY spectra obtained for A2rCrUr-labeled [5′-LΔPBS]2 and ΨCES confirm that the tandem three-way junction structure is present in both constructs. (E) NMR-derived secondary structure of ΨCES. Black and blue arrows denote A-H2 NOEs observable in ΨCES and fr-ΨCES samples, respectively; red arrows highlight NOEs shown in panel (C and D); thin arrows denote very long-range NOEs. (F) Packaging of native HIV-1NL4-3 5′-L and 5′-LΔPBS RNAs under competition conditions assayed by means of ribonuclease protection. P, undigested probe; M, RNA sizes marker. Lanes 1 and 2: native HIV-1NL4-3 helper versus test vectors containing 5′-LΔPBS (1) or native HIV-1NL4-3 (2). Lane 3: HIV-1NL4-3 helper expressed without test RNA. Lane 4: mock transfected-cells. Samples obtained from transfected cells (Cells) or viral containing media (Virus) are indicated. Bands corresponding to host 7SL RNA, HIV-1NL4-3 helper RNA (Ψ+) and co-packaged test RNAs (Test) are labeled.