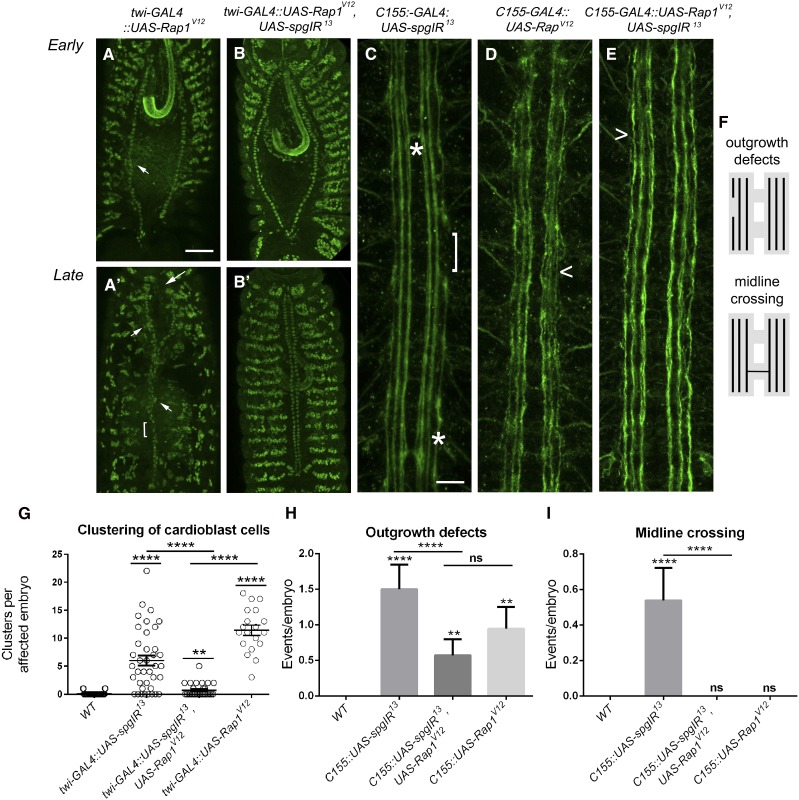
Figure 7.
Expression of RapV12 can temper spg−/− phenotypes. (A–E) Stage 13 (A and B) or stage 17 (A′, B′, C, and D) embryos labeled with α-mef2 to visualize muscle nuclei (A–B′) or α-1D4 (C–E) to label the longitudinal axons of the CNS. (A and A′) Embryos overexpressing Rap1V12 show early (A) and late (A′) clustering defects (A and A′, arrowheads), abnormal posterior openings (A, arrows), and unpaired cardioblasts (A′, bracket). (B and B′) The clustering defects observed upon loss of spg using RNAi (Figure 3, E and E′) are ameliorated with co-expression of the constitutively active form of Rap. (C) RNAi knockdown of spg (UAS-spgIR13) using the pan neuronal driver, C155-GAL4, gives rise to outgrowth defects (bracket) and midline crossovers (asterisks). (D) Overexpression of RapV12 on its own does not show guidance errors, but instead results in minor unbundling of the longitudinal axons (D, caret). (E) Outgrowth and guidance defects seen when knocking down Spg protein levels by RNAi (C, asterisks and bracket) are tempered when simultaneously expressing RapV12. (F) Schematic of outgrowth and midline crossing defects observed in spg RNAi embryos. (G–I) Graphs showing the ability of UAS-RapV12 to suppress cardioblast clustering (G), CNS outgrowth defects (H), or guidance errors due to inappropriate midline crossing (G) in a spg RNAi background. P-values are results of Mann–Whitney comparisons (G) of the bars indicated or Kruskal–Wallis tests (H and I) compared to wild type or the bars indicated (****P < 0.0001). Mean ± SEM. Posterior is up for all embryos. Bar: 50 μm (A–B′) and 10 μm (C–E).