Figure 5.
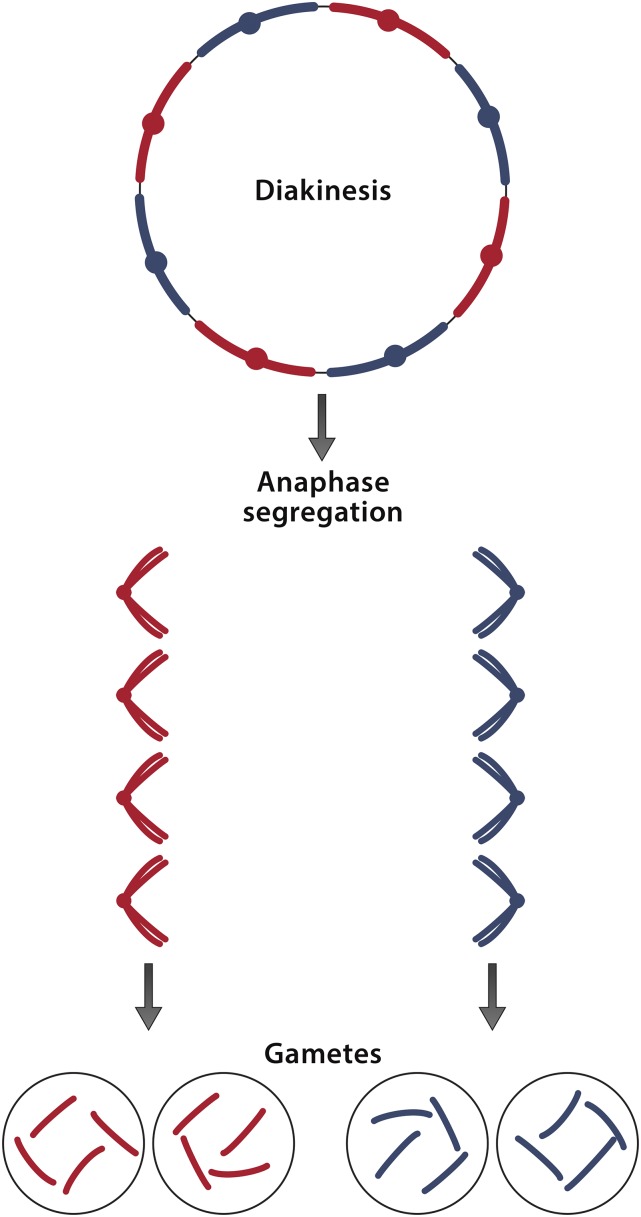
Oenothera-like meiosis depicted in a diploid genome of eight chromosomes. At diakinesis, all eight chromosomes are found joined end to end, forming a ring in which chromosomes derived from one parent (red) alternate with chromosomes derived from the other parent (blue). The two groups of chromosomes are referred to as complexes. There is no crossing over between complexes except possibly in telomeric regions where genetic exchange occasionally occurs. At first anaphase, chromosomes belonging to different complexes segregate to opposite poles. Entire haploid genomes therefore pass through meiosis intact, generation after generation. Rearrangements within a complex erode chromosome structural homology but do not change the genetic content of the complex in which they occur. Segregation is therefore accomplished without requiring homologous chromosome pairs. Oenothera-like meiosis in the present case would suggest that the genomes of isolates MA, MM, and CR are each composed of two complexes and that each complex includes an allele of each of the four sequenced regions. Owing to the identity in each region of MA1 with MM1 and the near identity of MA2 with CR2, there would be four distinct complexes represented in the three isolates: MA1/MM1, MA2/CR2, MM2, and CR1. Designating these complexes as 1, 2, 3, and 4, the genomes of MA, MM, and CR would be 1/2, 1/3, and 4/2, respectively. The multigeneration stability of each complex would then account for the apparent lack of assortment among the sequenced regions of isolates MA, MM, and CR.