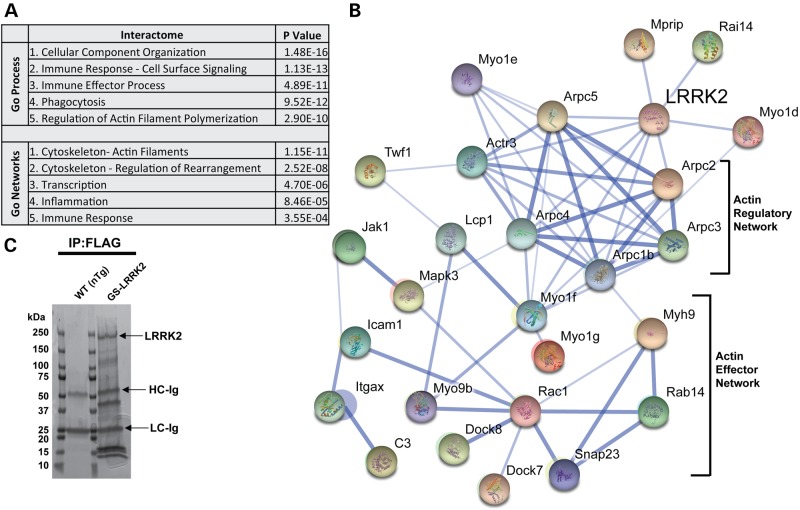
Figure 5.
Definition of the LRRK2 interactome in macrophages. (A) Gene-ontology (GO) process and network analysis to detect enrichment of terms from proteins identified in LRRK2-positive protein complexes (see Supplementary Material, Table S1 post-filter and Methods). P-values were calculated from Wilcoxon signed rank tests and corrected for multiple testing by Benjamini–Hochberg analysis. (B) String-DB analysis of LRRK2 proteins in complex that segregate into the actin-regulatory network and actin-cytoskeleton effector proteins, known to control myeloid cell motility. (C) Representative Coomassie-stained gel of immunoprecipitated proteins from M1-activated (LPS treated, 100 ng ml−1) TEPM from WT (non-transgenic, nTg) and G2019S(GS)-LRRK2 mice. Immunoprecipitates on beads were washed in parallel between nTg and G2019S LRRK2 FLAG-pull-downs so that the only protein bands visible in the nTg pull-down were derivative from the beads (i.e. FLAG antibody). Labeled bands include LRRK2 protein, and the heavy-chain and light-chain from the FLAG resin (HC and LC, respectively).