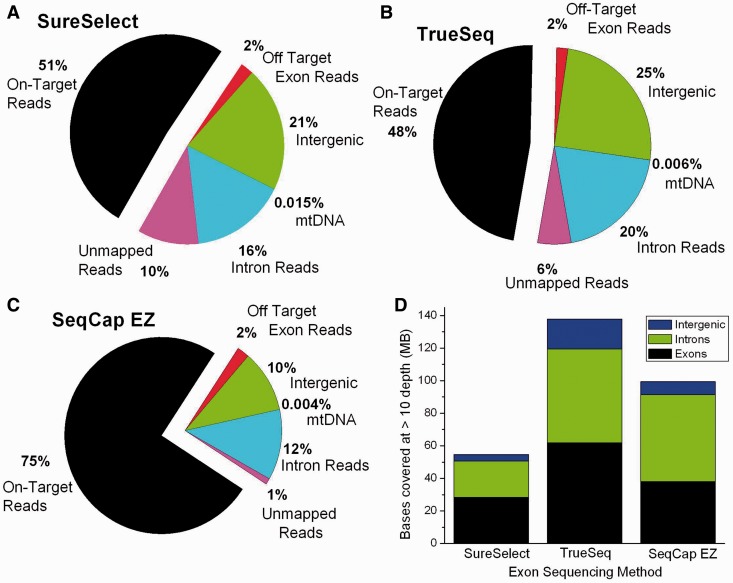
Figure 1:
The percentages of reads assigned to different categories for (A) SureSelect (v2), (B) TrueSeq and (C) the SeqCap EZ methods of exome sequencing. In all cases, the largest category of reads consists of the targeted genomic regions, but a large fraction of the reads are off target. The categories shown are the reads that map to exons that were not part of the target set, intergenic regions, mtDNA, introns and finally reads that do not map to any part of the human reference sequence. (D) The total number of bases covered at >10 depth that map to exons (both targeted and non-targeted exons), introns and intergenic regions for three methods of exome sequencing. These numbers should be compared with the full human genome size of approximately 3 billion base pairs.