Abstract
More than 30 years of research on nuclear RNA polymerases (RNAP I, II, and III) has uncovered numerous factors that regulate the activity of these enzymes during the transcription reaction. However, very little is known about the machinery that regulates the fate of RNAPs before or after transcription. In particular, the mechanisms of biogenesis of the 3 nuclear RNAPs, which comprise both common and specific subunits, remains mostly uncharacterized and the proteins involved are yet to be discovered. Using protein affinity purification coupled to mass spectrometry (AP–MS), we recently unraveled a high-density interaction network formed by nuclear RNAP subunits from the soluble fraction of human cell extracts. Validation of the dataset using a machine learning approach trained to minimize the rate of false positives and false negatives yielded a high-confidence dataset and uncovered novel interactors that regulate the RNAP II transcription machinery, including a set of proteins we named the RNAP II-associated proteins (RPAPs). One of the RPAPs, RPAP3, is part of an 11-subunit complex we termed the RPAP3/R2TP/prefoldin-like complex. Here, we review the literature on the subunits of this complex, which points to a role in nuclear RNAP biogenesis.
Keywords: RNA polymerase, biogenesis, protein affinity purification, RNA polymerase II-associated proteins (RPAPs), transcription
Introduction
Nuclear gene expression in eukaryotes is orchestrated by 3 RNA polymerases (RNAPs) that synthesize RNA molecules from a DNA template. RNAP I generates most ribosomal RNAs (rRNA), RNAP II synthesizes protein-coding mRNAs (mRNA), along with non-coding micro RNAs (miRNA), small nuclear RNAs (snRNA), and small nucleolar RNAs (snoRNA), and RNAP III mostly transcribes transfer RNAs (tRNA) and 5S rRNAs. Over the past 3 decades, many efforts have been made to identify and characterize the myriad protein factors that regulate the activity of nuclear RNAPs during the act of transcription. They include the DNA-binding transcriptional regulators (Tjian and Maniatis 1994; Triezenberg 1995; Ptashne and Gann 1997; Carey 1998), the general transcription factors (Orphanides et al. 1996; Conaway and Conaway 1997; Hampsey 1998; Coulombe and Burton 1999), and the co-activators or co-repressors (Conaway et al. 2005; Kornberg 2005; Roeder 2005; Marr et al. 2006). Studying the role of the RNAP co-regulators has revealed the existence of a multitude of proteins that participate in transcriptional regulation by acting on the organization or chemical modification of chromatin, the template of RNAPs in eukaryotic cells (Kornberg and Lorch 1999; Orphanides and Reinberg 2000; Li et al. 2007).
Surprisingly, and despite many efforts to analyze the regulatory mechanisms targeting transcription and transcription factors themselves, very little is known about the machinery that regulates nuclear RNAPs before or after transcription. For example, the biogenesis of RNAPs, which comprise both common and specific subunits, remains uncharacterized. This lack of information leaves many important questions unanswered. Are the different nuclear RNAPs assembled in the cytoplasm before being imported to the nucleus? Are they assembled as sub-complexes first, that are then assembled into active enzymes? Is a common mechanism in place or is there a distinct mechanism for each enzyme? Finding answers to these questions would be facilitated by the identification and characterization of regulatory factors involved in RNAP subunit folding, their assembly into active enzymes, and their nuclear import. Although RNAP biogenesis has been the object of a few reports (Hardeland and Hurt 2006), a molecular description of the process, including the identification and characterization of the regulatory factors involved, is still awaited.
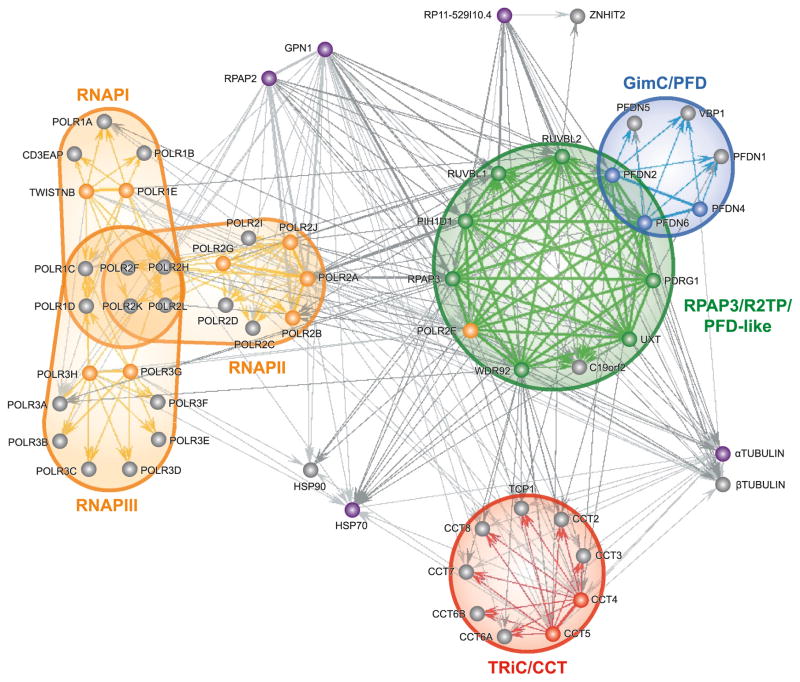
Because protein complexes involved in the assembly, folding, and nuclear import of RNAPs are likely found in the soluble fraction of cell extracts, as opposed to the insoluble fraction that contains chromatin and actively transcribing RNAP molecules, we surveyed soluble protein complexes that associate with RNAP subunits using protein affinity purification coupled to mass spectrometry (AP–MS). Specific subunits of all 3 nuclear RNAPs were tagged and purified to identify associating partners by MS. High confidence interactions were computationally inferred and most were confirmed by reciprocal tagging and purification. The resulting dataset stemming from 77 tagged proteins was used to draw a map of interactions connecting these various complexes (Cloutier et al. 2009). The composition and organization of this network revealed important features of the machinery involved in eukaryotic transcription. Notably, we identified a set of proteins that are tightly connected to RNAP II. Accordingly, these proteins were named RNAP II-associated proteins (RPAPs). Although the exact function of these factors remains elusive, one of these RPAPs, RPAP3, is part of an 11-subunit protein complex that we termed the RPAP3/R2TP/prefoldin-like complex (Fig. 1) based on similarity to the previously characterized R2TP complex, a cofactor of Hsp90 (Zhao et al. 2008), and prefoldin complexes (chaperone associated with the CCT/TRiC complex) (Geissler et al. 1998; Vainberg et al. 1998).
Fig. 1.
Diagram of the network of high-confidence interactions formed around the RPAP3/R2TP/prefoldin-like complex. Nodes represent tagged subunits of the RPAP3/R2TP/prefoldin-like complex (RPAP3/R2TP/PFD-like), classical prefoldin complex (GimC/PFD), CCT chaperone (TRiC/CCT), and RNA polymerases I, II, and III (RNAP I, RNAP II, RNAP III). Tagged proteins that have not been associated to any known complex are also indicated. Thin arrows represent unidirectional interactions, whereas thicker lines symbolize bidirectional interactions as detected in reciprocal tagging experiments. Lines and arrows of the same shade show interactions within a same complex (note that the term interaction does not signify direct binding between two proteins, but rather copurification of a tagged bait protein with its prey or targets. For simplification purposes, many gene isoforms have been condensed into one node: HSP90 (HSP90AA1, HSP90AA2, HSP90AB1), HSP70 (HSPA1A, HSPA1B, HSPA1L, HSPA2, HSPA5, HSPA6, HSPA8, HSPA9), αTUBULIN (TUBA1A, TUBA1B, TU-BA1C, TUBA3C, TUBA3D, TUBA3E, TUBA4A, TUBA4B, TUBA6), βTUBULIN (TUBB, TUBB1, TUBB2A, TUBB2B, TUBB2C, TUBB3, TUBB4, TUBB4Q, TUBB6). The tagged isoforms of HSP70 and αTUBULIN are HSPA8 and TUBA1A, respectively. (Note: a full-colour version of this image is available from http://bcb.nrc.ca.)
Similar complexes have been described in two independent reports. The first report described an approximately 1 MDa complex composed of Tip49 (RUVBL1), Tip48 (RUVBL2), PFD2 (PFDN2), PFDN4r (PDRG1), STAP1 (UXT), URI (C19orf2), and RPB5 (POLR2E) (Gstaiger et al. 2003). Whereas this report failed to detect the remaining 4 subunits of RPAP3/R2TP/prefoldin-like complex, we noted that a number of unidentified protein bands on the SDS gel could be attributed to the missing components. Based on their approximated molecular masses, p10, p40, and p42 could be PFDN6, PIH1D1 and WDR92, respectively, whereas RPAP3 could be present in the upper portion of the large 90 kDa band where C19orf2 was identified.
The other paper to describe this complex was much more detailed and identified all of the same components, but included the RNAP III catalytic subunit POLR3A/RPC1 (Sardiu et al. 2008). Our own assays have identified POLR3A on occasion, but based on the weak mascot score attributed to this protein (which can also be found in the heatmap representation of Sardiu et al. (2008)), we propose this reflects a transient interaction of the RPAP3/R2TP/prefoldin-like complex with RNAP III. Moreover, once the central components of the RPAP3/R2TP/prefoldin-like complex were defined, we were able to observe association with subunits of all 3 nuclear RNAPs. How exactly this complex relates to RNAP function is still under investigation.
RPAP2 and GPN1/RPAP4 are additional interactors that we have found to be tightly connected to RNAPs and the RPAP3/R2TP/prefoldin-like complex. The yeast homologue of RPAP2, Rtr1, has been shown to affect transcription by RNAP II and was identified as an RNAP II specific phophatase that targets the COOH-terminal domain (CTD) of the largest subunit POLR2A/RPB1 during transcription, bringing about a change in the phosphorylation pattern of the CTD that is required for recruitment of termination factors (Gibney et al. 2008; Mosley et al. 2009). GPN1 is a GTPase that appears to have a role in RNAP II transport from the cytosol to the nucleus (Forget, D., Lacombe, A., and Coulombe, B., unpublished observations). Finally, it should come as no surprise that Hsp90 and the TRiC/CCT chaperones also appear in this network based on their well-known interaction with R2TP and the prefoldin complex, respectively (Vainberg et al. 1998; Zhao et al. 2005b).
The purpose of this review is to outline the literature that has been generated on all 11 subunits of the RPAP3/R2TP/prefoldin-like complex to define a general framework for the putative role for this complex in regulating RNAP. This review will also focus on a few other proteins shown to interact transiently or weakly with the complex, as they might also provide insights into its function and (or) regulation.
The R2TP module
RUVBL1 and RUVBL2 (RVB1 and RVB2; TIP49 and TIP48; pontin and reptin)
RUVBL1 and RUVBL2 are ATPases of the AAA+ superfamily (ATPase associated with diverse cellular activities) and are related to the prokaryotic DNA helicase RuvB (Qiu et al. 1998; Kanemaki et al. 1999), which is involved in homologous recombination and double-strand break repair (see Fig. 2 for a schematic representation of the R2TP module components). In metazoans, these proteins assemble into two distinct homohexameric rings that interact with one another to form a large dodecameric unit (Puri et al. 2007), or heterohexamer as is the case for the yeast homologs Rvb1 and Rvb2 (Gribun et al. 2008), which is an integral part of the Ino80, SRCAP, and TRRAP/TIP60 chromatin-modifying complexes (Sardiu et al. 2008). They are also required for ribonucleoprotein (RNP) assembly of snoRNPs (Newman et al. 2000; King et al. 2001) and telomerase (Venteicher et al. 2008).
Fig. 2.
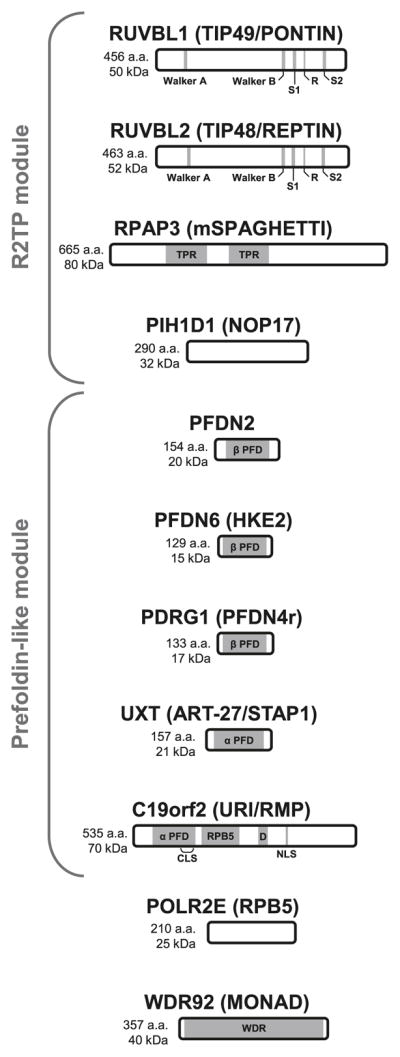
Schematic representation of the 11 subunits of the RPAP3/R2TP/prefoldin-like complex. Domain architecture, length, and molecular mass of proteins are shown. AAA+ ATPase motifs of RUVBL1 and RUVBL2 consist of Walker A and Walker B elements, 2 sensors (S1 and S2), and a protruding arginine finger (R) essential for hydrolysis. TPR, tetratricopeptide repeats; α PFD, alpha prefoldin homology domain; β PFD, beta prefoldin homology domain; RPB5, RPB5-binding domain; D, aspartic acid-rich region; CLS, cytoplasmic localization signal; NLS, nuclear localization signal; WDR, WD40 repeats.
Interestingly, RUVBL2 has been shown to be the target of sumoylation at ser139 (Kim et al. 2006). Mutation of this residue leads to cytoplasmic localization and derepression of the metastatic suppressor KAI1 by Ino80. On the other hand, when a chimerical version of RUVBL2 is expressed with the SUMO peptide present at the N-termini of the protein, it becomes exclusively nuclear and KAI1 repression is restored. Though the enzymes responsible for this modification have been identified as being the E2 SUMO-conjugating enzyme ubc9 and the desumoylating enzyme SENP1, there is no mention of a similar mechanism for RUVBL1, which bears close homology with RUVBL2.
RPAP3 (Spaghetti/Tah1)
In a previous paper, we had identified RPAP3 as a novel interactor of RNAP II (RNAP II-Associated Protein 3) (Jeronimo et al. 2007). This protein has 2 tetratricopeptide repeat (TPR) domains that have been shown to mediate interaction with Hsp90 in a similar fashion as another TPR-containing Hsp90 cofactor, Hop/Sti-1 (Scheufler et al. 2000; Boulon et al. 2008).
A role for RPAP3 in DNA repair and apoptosis has been proposed. In a recent article it was shown that overexpression of RPAP3 sensitizes HEK293 cells to UV-induced cell death and phosphorylation of histone variant H2AX, a marker of double strand breaks that targets repair factors to the site of DNA damage. Knockdown of RPAP3 by RNAi had an altogether opposed effect on every phenotype studied as did overexpression of the related protein RUVBL2, suggesting a possible inhibitory function (Ni et al. 2009).
It has also been reported that in Drosophila, P element-induced insertions in the gene of the RPAP3 homolog, Spaghetti, results in defects in the development of imaginal discs responsible for wing morphogenesis (Roch et al. 1998).
PIH1D1 (Pih1/Nop17)
The recent mapping of the physical and genetic interaction network of Saccharomyces cerevisiae Hsp90 led to the identification of a novel cofactor composed of Rvb1 and Rvb2, Tah1, and Pih1/Nop17. This cofactor complex was termed the R2TP (Rvb1–Rvb2–Tah1–Pih1) complex (Zhao et al. 2005b).
At the time the R2TP complex was characterized, Nop17 had already been shown to have a role in rRNA maturation. A yeast two-hybrid (Y2H) screen revealed this nucleolar protein interacts with exosome component Rrp43 (involved in rRNA processing), the box C/D snoRNP protein Nop58 and the presumed box H/AHA snoRNP assembly factor Nop53 (Gonzales et al. 2005; Granato et al. 2005). Deletion of the Nop17 gene led to dissociation from the nucleolus of all 4 box C/D snoRNP, Nop1, Nop56, Nop58, and snu13 and accumulation of pre-rRNA 35 S (Gonzales et al. 2005). Notably, Pih1/Nop17 is also an unstable protein with a propensity to aggregate in the absence of Tah1 and Hsp90 (Zhao et al. 2008).
Assuming that yeast Tah1 is the homolog of RPAP3/Spaghetti (despite a size discrepancy, both proteins share a high degree of homology and a conserved domain architecture), the ability of R2TP components to form a complex seems to be conserved in both Drosophila (Giot et al. 2003) and human (Te et al. 2007). Furthermore, study of the mammalian counterparts yielded similar results in that they associated with U3 snoRNA, but also U4 snRNP, and SBP2. It was therefore proposed that R2TP might be involved in maturation of not only box C/D snoRNPs, but possibly all L7Ae RNPs, of which box C/D snoRNPs are members, along with box H/ACA snoRNPs, U4 snRNP, telomerase, and selenoprotein-coding mRNPs. Inhibition of Hsp90 by geldanamycin seems to validate the assumption that Hsp90 and its co-chaperone R2TP might act during L7Ae RNP assembly as it resulted in a decrease in the number of L7Ae proteins (Boulon et al. 2008).
Our own AP–MS analysis of the R2TP subunits has not identified L7Ae protein family members. The reverse is also true, upon AP–MS analysis of the box C/D snoRNP proteins FBL (Nop1 homologue) and NOP56, no RPAP3 complex component was identified. Of note, another PIH domain containing protein exists in human, PIH1D2, though its function remains unknown and it has not appeared in any of our AP–MS experiments.
The prefoldin-like module
PFDN2 and PFDN6
The GimC/prefoldin (PFD) complex is a highly conserved heterohexameric chaperone composed of two classes of proteins: α and β prefoldins. Prefoldins of the β group (PFDN1, PFDN2, PFDN4, and PFDN6/HKE2) consist of two long α helices in a coiled-coil conformation connected by a β hairpin linker comprising two short β strands, whereas α prefoldins (VBP1/PFDN3, PFDN5) share the same arrangement but have two β hairpins instead of one (see Fig. 2 for a schematic representation of the prefoldin-like module components). Once assembled, the β hairpins form the core of the complex, two 8-stranded β barrels, while the coiled-coil helixes resemble tentacle-like appendages that first mediate substrate binding (Siegert et al. 2000; Lundin et al. 2004). The PFD complex binds unfolded, newly synthesized poly-peptides and delivers them to the CCT chaperone for proper folding. The best known targets of this prefoldin–CCT pathway are the cytoskeletal proteins actin and tubulin (Geissler et al. 1998; Vainberg et al. 1998; Siegers et al. 1999).
In addition to their involvement in the classical PFD complex, PFDN2 and PFDN6 were found in the RPAP3/R2TP/prefoldin-like complex. Of note, this novel complex differs from the classical PFD complex in that it only has 5 prefoldin or prefoldin-like proteins, but we cannot assume a pentameric conformation as one of these subunits might be present in two copies.
PDRG1 (PFDN4r)
Very little is known about PDRG1 other than the observation that its expression is repressed by the tumor suppressor p53. Ultraviolet radiation was shown to upregulate PDRG1 gene expression via binding of the Oct-1 transcription factor to a conserved element in its promoter region (hence the name, p53 and DNA damage-regulated gene 1). GFP-tagged fusions of this protein were shown to localize primarily in cytosolic aggregates, possibly due to association with some yet-to-be-identified subcellular compartment. Overexpression of PDRG1 further sensitized cells to UV irradiation, pointing to a possible role in apoptosis (Luo et al. 2003).
Though the authors mentioned not having found significant homology to any known protein, PDRG1 is most likely the PFDN4-related (PFDN4r) protein mentioned in Gstaiger et al. (2003), given the resemblance between these two proteins (about 25% identity and 53% similarity). Notably, in our own AP–MS experiments, PDRG1 was the only prefoldin-like protein to copurify other canonical prefoldins, unlike C19orf2 and UXT, which yielded only PFDN2 and PFDN6. Indeed, in addition to these two aforementioned prefoldins, we were able to detect PFDN1, VBP1/PFDN3 and PFDN5, but not PFDN4, which could imply that in addition to the prefoldin-like module of the RPAP3/R2TP/prefoldin-like complex, PDRG1 could be present in a third prefoldin complex, closer in structure and composition to the archetypical PFD where it would substitute for PFDN4. How would this alteration affect the specificity of the prefoldin complex is still unknown.
UXT (ART-27/STAP1)
UXT/ART-27 was mapped to the Xp11 locus, a region responsible of a number of genetic disorders, including Renpenning, Prieto, and Sutherland–Haan syndromes, but does not appear to be involved in their onset (Schroer et al. 1999).
It was shown to associate with the N-terminus of the androgen receptor (androgen receptor (AR) trapped clone 27 or ART-27) by a Y2H screen and enhance transcriptional activation by a number of steroid and thyroid receptors, including AR, the glucocorticoid receptor (GR), the estrogen receptor (ER)-α, ER-β, and the thyroid receptor (TR) β-1. This activity might be attributable to UXT alone, although the same report mentioned that this protein was part of a larger complex as observed by velocity gradient sedimentation of HeLa extracts (Markus et al. 2002).
This once considered “ubiquitously expressed transcript” is actually tightly regulated both in a cell line- and developmental stage-specific fashion and is downregulated in prostate cancer cells, consistent with a role as an inhibitor of androgen-mediated cellular proliferation (Taneja et al. 2004; Nwachukwu et al. 2007). In a separate study, UXT was also shown to bind the transcriptional repressor Evi1 and inhibit its cell transformation property in Rat1 cells (McGilvray et al. 2007). Conversely, UXT was found to be overexpressed in many other tumor cell lines (Schroer et al. 1999; Zhao et al. 2005a).
A different Y2H screen identified UXT as interacting with the centrosomal protein phosphatase Cdc14A. Immunofluorescence showed that UXT colocalizes with γ tubulin to the centrosome and that overexpression of this prefoldin-like protein results in a disruption of normal centrosomal structure. The authors of this paper assume that a resulting defect in chromosome segregation may account for the apparent tumorigenic nature of this protein (Zhao et al. 2005a).
Surprisingly, yet another series of reports stated that UXT interacted with a mitochondria-associated leucine-rich penta-tricopeptide repeat motif-containing (LRPPRC) protein and that expression of a GFP-tagged UXT exhibited differential localization based on its expression level. At lower concentration, UXT appears to be diffusely cytoplasmic and gradually aggregates in a perinuclear pattern along with mitochondria, a hallmark of some forms of cell death. Higher levels of GFP–UXT expression result in nuclear invasion and subsequent disruption of the nucleus. An additional interaction with the mitotic checkpoint protein BUB3 was reported, but no apparent colocalization was observed (Liu and McKeehan 2002; Moss et al. 2007).
C19orf2 (URI/RMP)
URI/RMP is likely the most intensively studied member of prefoldin-like family and its large size has garnered it the name unconventional prefoldin RPB5 interactor (URI). This extended C-terminal structure is responsible for direct binding of the nuclear RNAP common subunit RPB5 (hence the name RPB5 mediating protein, or RMP).
URI/RMP appears to have a corepressor activity, dependent on the general transcription factor TFIIF with which both RMP and RPB5 interact on transactivation via the hepatitis B virus X protein (HBx). This activity would proceed through a competition for binding of the RPB5 subunit (Nikolov and Burley 1997; Dorjsuren et al. 1998; Wei et al. 2003), consistent with the proposed role of RPB5 on activated transcription (Miyao and Woychik 1998).
URI/RMP is mainly located in the cytoplasm, but does contain a nuclear localization signal and can translocate to the nucleus upon DNA methyltransferase 1-associating protein (DMAP1) overexpression (Delgermaa et al. 2004). Mutants in Drosophila melanogaster and Caenorhabditis elegans have both shown an accumulation of DNA damage, suggesting that whatever fraction of URI/RMP that locates to the nucleus might have a role in chromatin stability (Parusel et al. 2006; Kirchner et al. 2008).
More recently, it has also been shown that URI/RMP is phosphorylated by S6K1 and acts as a regulator of gene expression in the nutrient-sensitive TOR signaling pathway —an activity it shares with its yeast counterpart, BUD27 (Gstaiger et al. 2003). Moreover, URI/RMP phosphorylation leads to the release of the phosphatase PP1γ from its tethered state at the surface of mitochondria and subsequently acts in a negative feedback loop to alleviate the effect of S6K1-mediated phosphorylation of the apoptotic protein BAD (Djouder et al. 2007). This ability of URI/RMP to bind PP1α-type phosphatases, of which human PP1γ is a member, is conserved even in Drosophila (Kirchner et al. 2008).
A role of URI/RMP in protein translation, more precisely by coupling translation initiation and quality control, has also been proposed. Indeed, BUD27 has also been shown to form a complex with the cotranslational quality control chaperones Ssb1p and Sis1p, as well as the elongation factor eEF1A, and to be somehow involved with initiating translation in yeast (Deplazes et al. 2009).
Other RPAP3/R2TP/prefoldin-like complex components
POLR2E (RPB5)
As mentioned previously, RPB5 (POLR2E) is a subunit shared by all 3 nuclear RNAPs (see Fig. 2 for a schematic representation of the other components of the RPAP3/R2TP/prefoldin-like complex). Our own results indicate that RPB5 is not merely the target by which the RPAP3/R2TP/prefoldin-like complex interacts with RNAPs, but should instead be considered as a bona fide component of this new complex. Of note, though it might be easy to assume that the presumed chaperone activity of the RPAP3/R2TP/prefoldin-like complex might be directed towards ensuring proper folding of RPB5 prior to its association with RNAP holoenzymes, the fact that the RPB5-binding domain of URI/RMP is separate from its prefoldin homology domain may preclude RPB5 from being the target of prefoldin-CCT mediated folding.
WDR92 (MONAD)
Sequence analysis of WDR92 (Monad) predicts that the bulk of the protein is folded into a β-propeller. This unique symmetrical structure is made up of several WD40 repeats (seven in WDR92 as opposed to the two reported by Saeki et al. (2006) that each consists of a small four-stranded anti-parallel β-sheet. The resulting platform may be a mediator of the interaction between the R2TP and prefoldin-like modules.
WDR92 has been reported to potentiate TNFα and cycloheximide-induced apoptosis in a manner similar to its interacting protein RPAP3 (Saeki et al. 2006; Itsuki et al. 2008). Alternatively, WDR92 could be a target of the presumed chaperone activity of the RPAP3/R2TP/prefoldin-like complex. Indeed, it has been shown that WD40 domains require assisted folding by the chaperones CCT and Hsp70 (Siegers et al. 2003). But given that no other WD40 domain-containing protein were as systematically identified as WDR92, this hypothesis of a WDR92-specific chaperone appears unlikely.
Interactors
Over the years, a number of reports have identified components of the RPAP3/R2TP/prefoldin-like complex as interactors of proteins that were not identified in the most recent AP–MS experiments describing the RPAP3/R2TP/prefoldin-like complex. These proteins warrant consideration here. The mechanism by which they affect the activity of the complex will undoubtedly be an interesting avenue of investigation.
PARD6A (Par-6C)
PAR proteins are asymmetrically localized factors required for the establishment of the anterior–posterior polarity axis early in Caenorhabditis elegans embryogenesis (Kemphues et al. 1988; Watts et al. 1996) (see Fig. 3 for a schematic representation of the interactors of the RPAP3/R2TP/prefoldin-like complex). Brajenovic and colleagues have undertaken the mapping of the interaction network of human orthologues of Par proteins by AP (Brajenovic et al. 2003). Surprisingly, one of these proteins, Par-6C, has yielded a number of RPAP3/R2TP/prefoldin-like complex subunits, including RPAP3 itself (mSpaghetti), WDR92 (FLJ31741), PIH1D1 (FLJ20643), and PDRG1 (C20orf126). Another PAR protein, Par-4, was copurified with RPAP3, but no other component of the complex was detected in that eluate nor any of the other Par-6 proteins (A and B) or typical Par-6 interactors, Par-3 and aPKC.
Fig. 3.
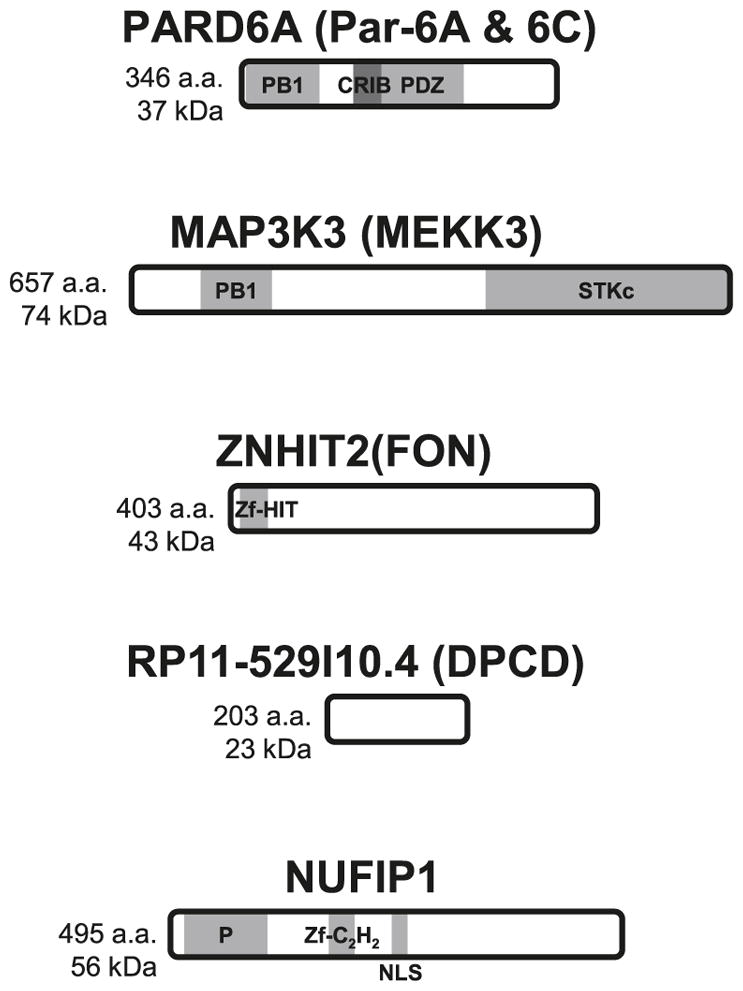
Schematic representation of five reported interactors of the RPAP3/R2TP/prefoldin-like complex. Domain architecture, length, and molecular mass of proteins are shown. PB1, Phox and Bem1p homology domain; CRIB, Cdc42/Rac interactive binding; PDZ, PSD-95, DlgA and ZO-1 homology domain; STKc, serine–threonine kinase catalytic domain; Zf-HIT, HIT-type zinc finger motif; Zf-C2H2, C2H2-type zinc finger motif; P, proline-rich region; NLS, nuclear localization signal.
MAP3K3 (MEKK3)
MEKK3 is a member of the mitogen-activated protein kinase kinase kinase (MAP(3)K) family. MAP(3)Ks participate in the early steps of a signal transduction pathway that will promote various cellular processes in responses to a number of external stimuli through a sequential MAP(3)K > MAP(2)K > MAPK phosphorylation cascade (Chang and Karin 2001; Pearson et al. 2001). In accordance to this general scheme, MEKK3 is known to phosphorylate a number of MAP(2)K (MEK/MKK1–7), which leads to the activation of their specific downstream MAPK (ERK1/2, JNK/SAPK, ERK5, and p38) (Blank et al. 1996; Deacon and Blank 1997; Ellinger-Ziegelbauer et al. 1997; Chao et al. 1999; Deacon and Blank 1999; Uhlik et al. 2003). MEKK3 is also involved in the NF-κB pathway where it activates the IKK complex in response to TNFα and LPS (Yang et al. 2001; Schmidt et al. 2003; Samanta et al. 2004).
In a landmark report aimed at mapping the entirety of the interaction network of the TNF-α/NF-κB signal transduction pathway by AP–MS, MEKK3 was found in association with the majority of RPAP3/R2TP/prefoldin-like complex subunits (Bouwmeester et al. 2004). Indeed, RPAP3 (FLJ21908), PIH1D1 (FLJ20643), RUVBL1, RUVBL2, PFDN2, PFDN6 (HKE2), PDRG1 (C20orf126) and C19orf2 all copurified with MEKK3, as did the CCT complex and Hsp90α/β. Of note, other classical prefoldins were present, including PFDN1, VBP1 and PFDN4, which may suggest that MEKK3 association might be mediated by the prefoldin-like module.
There might be a link between these observations and a recent paper that states that UXT is part of the NF-κB enhanceosome (Sun et al. 2007). Though the authors favored a model where UXT interacts directly with the p65 subunit of NF-κB, many of the results (NF-κB-dependent transcription, subcellular NF-κB localization, TNFα-mediated apoptosis) might be explained by a RPAP3/R2TP/prefoldin-like complex-dependent regulation of the activity of upstream MEKK3 activator.
ZNHIT2 and RP11–529I10.4
ZNHIT2 and RP11–529I10.4 were found initially in both our RUVBL1 and RUVBL2 affinity purifications. To our surprise, reciprocal tagging of the RP11–529I10.4 protein revealed not only the presence of the RUVBL1 and RUVBL2 subcomplex, but the entire RPAP3/R2TP/prefoldin-like complex. Once again, these data are reminiscent to those presented in Sardiu et al. (2008). In addition, ZNHIT2 was also present in RP11–529I10.4 purification. Though we cannot speculate on a possible heterodimer confirmation between these two proteins, it seems they interact together with the RPAP3/R2TP/prefoldin-like complex, most likely through the RUVBL1 and RUVBL2 components. Very little is known about both factors.
ZNHIT2/FON is part of a family of proteins that contain a novel zinc-binding moiety called the zinc finger Hit domain (zf-HIT) that binds two zinc atoms in a core composed of two antiparallel β sheets followed by a short α helix, a conformation that is reminiscent of the B-box, RING and PHD domains (He et al. 2007). Other zf-HIT domain-containing proteins include ZNHIT1 of the SRCAP chromatin remodeling complex (Sardiu et al. 2008) and ZNHIT3/TRIP3, a coactivator of the HNF4α transcription factor (Iwahashi et al. 2002).
Deletion of RP11–529I10.4/DPCD in mice results in a phenotype similar to primary ciliary dyskinesia (PCD) (Zariwala et al. 2004). PCD is a genetic disorder characterized by a defect in axonemal structure resulting in hindrance of normal ciliated and flagellar cells, most notably of the respiratory tract. Whether or not this phenotype has anything to do with a possible microtubular polymerizing activity of the RPAP3/R2TP/prefoldin-like complex is an interesting hypothesis that remains to be investigated.
NUFIP1
NUFIP is another zinc finger motif-containing protein that was first identified by Y2H screens as a FMRP (FMR1) interactor (Bardoni et al. 1999). This shuttling protein was later found to interact also with BRCA1 and the Cyclin T1 subunit of P-TEFb and promote RNAP II transcription and ATP-dependent disassembly of hyperphosphorylated form of the holoenzyme from open transcription complexes (Cabart et al. 2004).
In the network built by Sardiu and colleagues, NUFIP is shown to be a RPAP3/R2TP/prefoldin-like complex interactor (Sardiu et al. 2008), which is not surprising given that both NUFIP and its yeast homologue, RSA1, where able to bind the R2TP components PIH1D1 (PIH1/Nop17), RUVBL1 (TIP49) and RUVBL2 (TIP48) (McKeegan et al. 2007; Boulon et al. 2008). In both studies a role for NUFIP and their associated factors in box C/D snoRNP (or even all L7Ae family RNP) biogenesis was proposed.
Though the exact purpose of this interaction is, again, not clear, it was noted that immunodepletion assay followed by add-back of recombinant NUFIP was not sufficient to restore RNAP II transcription in vitro unlike the addition of immunopurified NUFIP, which retains a number of essential cofactors (Cabart et al. 2004). It would therefore be interesting to assess whether the presence of the RPAP3/R2TP/prefoldin-like complex could be responsible for this effect.
Concluding remarks
AP–MS coupled to efficient computational biology methods offers a powerful procedure for the identification and characterization of protein complexes and networks. We used this procedure to unravel a novel protein complex that is tightly connected to nuclear RNAP subunits. This complex termed the RPAP3/R2TP/prefoldin-like complex is composed of proteins previously defined to have a role in protein complex assembly, such as some chaperone cofactors and the prefoldins. This finding points to a role of the RPAP3/R2TP/prefoldin-like complex at some stage of the process of nuclear RNAP biogenesis. One could speculate that RPB5 (POLR2E) might be a target of this presumed chaperone activity. On the other hand, RPB5 might only act as an anchor through which the RPAP3/R2TP/prefoldin-like complex could target other subunits of RNAPs, most notably the largest, catalytic subunits specific to each polymerase. Alternatively, the complex might have an activity that is closer in nature to the canonical prefoldin complex in that it could be involved in cytoskeletal assembly. Given that hyperphosphorylated RNAP II is known to migrate away from the DNA during mitosis (Parsons and Spencer 1997; Kim et al. 1997), association with RNAPs might therefore be a trigger to promote a cell-cycle dependent microtubular rearrangement or more simply, it could be responsible for directly segregating the RNA polymerases from DNA during this timeframe. Whatever the case might be, reviewing the literature on the many subunits and interactors of this complex, as we have done here, helps defining its role in RNAP biogenesis and its putative coordination with other cellular processes. This review will likely be the starting point of more focused and intensive efforts aimed at characterizing this important protein interaction network.
Acknowledgments
We wish to thank people of our laboratory for helpful discussions. This work was supported by grants from the Canadian Institutes for Health Research (CIHR) and the National Science and Engineering Research Council of Canada (NSERC).
Footnotes
This paper is one of a selection of papers published in this special issue entitled “Canadian Society of Biochemistry, Molecular & Cellular Biology 52nd Annual Meeting — Protein Folding: Principles and Diseases” and has undergone the Journal’s usual peer review process.
References
- Bardoni B, Schenck A, Mandel JL. A novel RNA-binding nuclear protein that interacts with the fragile X mental retardation (FMR1) protein. Hum Mol Genet. 1999;8(13):2557–2566. doi: 10.1093/hmg/8.13.2557. [DOI] [PubMed] [Google Scholar]
- Blank JL, Gerwins P, Elliott EM, Sather S, Johnson GL. Molecular cloning of mitogen-activated protein/ERK kinase kinases (MEKK) 2 and 3. Regulation of sequential phosphorylation pathways involving mitogen-activated protein kinase and c-Jun kinase. J Biol Chem. 1996;271(10):5361–5368. doi: 10.1074/jbc.271.10.5361. [DOI] [PubMed] [Google Scholar]
- Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jády BE, et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J Cell Biol. 2008;180(3):579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6(2):97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- Brajenovic M, Joberty G, Küster B, Bouwmeester T, Drewes G. Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J Biol Chem. 2003;279(13):12804–12811. doi: 10.1074/jbc.M312171200. [DOI] [PubMed] [Google Scholar]
- Čabart P, Chew HK, Murphy S. BRCA1 cooperates with NUFIP and P-TEFb to activate transcription by RNA polymerase II. Oncogene. 2004;23(31):5316–5329. doi: 10.1038/sj.onc.1207684. [DOI] [PubMed] [Google Scholar]
- Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92(1):5–8. doi: 10.1016/S0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410(6824):37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Chao TH, Hayashi M, Tapping RI, Kato Y, Lee JD. MEKK3 directly regulates MEK5 activity as part of the big mitogen-activated protein kinase 1 (BMK1) signaling pathway. J Biol Chem. 1999;274(51):36035–36038. doi: 10.1074/jbc.274.51.36035. [DOI] [PubMed] [Google Scholar]
- Cloutier P, Al-Khoury R, Lavallée-Adam M, Faubert D, Jiang H, Poitras C, et al. High-resolution mapping of the protein interaction network for the human transcription machinery and affinity purification of RNA polymerase II-associated complexes. Methods. 2009;48(4):381–386. doi: 10.1016/j.ymeth.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. General transcription factors for RNA polymerase II. Prog Nucleic Acid Res Mol Biol. 1997;56:327–346. doi: 10.1016/S0079-6603(08)61009-0. [DOI] [PubMed] [Google Scholar]
- Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30(5):250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Coulombe B, Burton ZF. DNA bending and wrapping around RNA polymerase: a “revolutionary” model describing transcriptional mechanisms. Microbiol Mol Biol Rev. 1999;63(2):457–478. doi: 10.1128/mmbr.63.2.457-478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon K, Blank JL. Characterization of the mitogen-activated protein kinase kinase 4 (MKK4)/c-Jun NH2-terminal kinase 1 and MKK3/p38 pathways regulated by MEK kinases 2 and 3. MEK kinase 3 activates MKK3 but does not cause activation of p38 kinase in vivo. J Biol Chem. 1997;272(22):14489–14496. doi: 10.1074/jbc.272.22.14489. [DOI] [PubMed] [Google Scholar]
- Deacon K, Blank JL. MEK kinase 3 directly activates MKK6 and MKK7, specific activators of the p38 and c-Jun NH2-terminal kinases. J Biol Chem. 1999;274(23):16604–16610. doi: 10.1074/jbc.274.23.16604. [DOI] [PubMed] [Google Scholar]
- Delgermaa L, Hayashi N, Dorjsuren D, Nomura T, Thuy TT, Murakami S. Subcellular localization of RPB5-mediating protein and its putative functional partner. Mol Cell Biol. 2004;24(19):8556–8566. doi: 10.1128/MCB.24.19.8556-8566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplazes A, Möckli N, Luke B, Auerbach D, Peter M. Yeast Uri1p promotes translation initiation and may provide a link to cotranslational quality control. EMBO J. 2009;28(10):1429–1441. doi: 10.1038/emboj.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djouder N, Metzler SC, Schmidt A, Wirbelauer C, Gstaiger M, Aebersold R, et al. S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol Cell. 2007;28(1):28–40. doi: 10.1016/j.molcel.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Dorjsuren D, Lin Y, Wei W, Yamashita T, Nomura T, Hayashi N, Murakami S. RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Mol Cell Biol. 1998;18(12):7546–7555. doi: 10.1128/mcb.18.12.7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger-Ziegelbauer H, Brown K, Kelly K, Siebenlist U. Direct activation of the stress-activated protein kinase (SAPK) and extracellular signal-regulated protein kinase (ERK) pathways by an inducible mitogen-activated protein Kinase/ERK kinase kinase 3 (MEKK) derivative. J Biol Chem. 1997;272(5):2668–2674. doi: 10.1074/jbc.272.5.2668. [DOI] [PubMed] [Google Scholar]
- Geissler S, Siegers K, Schiebel E. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J. 1998;17(4):952–966. doi: 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibney PA, Fries T, Bailer SM, Morano KA. Rtr1 is the Saccharomyces cerevisiae homolog of a novel family of RNA polymerase II-binding proteins. Eukaryot Cell. 2008;7(6):938–948. doi: 10.1128/EC.00042-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302(5651):1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Gonzales FA, Zanchin NI, Luz JS, Oliveira CC. Characterization of Saccharomyces cerevisiae Nop17p, a novel Nop58p-interacting protein that is involved in Pre-rRNA processing. J Mol Biol. 2005;346(2):437–455. doi: 10.1016/j.jmb.2004.11.071. [DOI] [PubMed] [Google Scholar]
- Granato DC, Gonzales FA, Luz JS, Cassiola F, Machado-Santelli GM, Oliveira CC. Nop53p, an essential nucleolar protein that interacts with Nop17p and Nip7p, is required for pre-rRNA processing in Saccharomyces cerevisiae. FEBS J. 2005;272(17):4450–4463. doi: 10.1111/j.1742-4658.2005.04861.x. [DOI] [PubMed] [Google Scholar]
- Gribun A, Cheung KL, Huen J, Ortega J, Houry WA. Yeast Rvb1 and Rvb2 are ATP-dependent DNA helicases that form a heterohexameric complex. J Mol Biol. 2008;376(5):1320–1333. doi: 10.1016/j.jmb.2007.12.049. [DOI] [PubMed] [Google Scholar]
- Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M, et al. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 2003;302(5648):1208–1212. doi: 10.1126/science.1088401. [DOI] [PubMed] [Google Scholar]
- Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62(2):465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeland U, Hurt E. Coordinated nuclear import of RNA polymerase III subunits. Traffic. 2006;7(4):465–473. doi: 10.1111/j.1600-0854.2006.00399.x. [DOI] [PubMed] [Google Scholar]
- He F, Umehara T, Tsuda K, Inoue M, Kigawa T, Matsuda T, et al. Solution structure of the zinc finger HIT domain in protein FON. Protein Sci. 2007;16(8):1577–1587. doi: 10.1110/ps.062635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsuki Y, Saeki M, Nakahara H, Egusa H, Irie Y, Terao Y, et al. Molecular cloning of novel Monad binding protein containing tetratricopeptide repeat domains. FEBS Lett. 2008;582(16):2365–2370. doi: 10.1016/j.febslet.2008.05.041. [DOI] [PubMed] [Google Scholar]
- Iwahashi H, Yamagata K, Yoshiuchi I, Terasaki J, Yang Q, Fukui K, et al. Thyroid hormone receptor interacting protein 3 (trip3) is a novel coactivator of hepatocyte nuclear factor-4alpha. Diabetes. 2002;51(4):910–914. doi: 10.2337/diabetes.51.4.910. [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27(2):262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Kurokawa Y, Matsuura T, Makino Y, Masani A, Okazaki K, et al. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J Biol Chem. 1999;274(32):22437–22444. doi: 10.1074/jbc.274.32.22437. [DOI] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52(3):311–320. doi: 10.1016/S0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kim E, Du L, Bregman DB, Warren SL. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136(1):19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Choi HJ, Kim B, Kim MH, Lee JM, Kim IS, et al. Roles of sumoylation of a reptin chromatin-remodelling complex in cancer metastasis. Nat Cell Biol. 2006;8(6):631–639. doi: 10.1038/ncb1415. [DOI] [PubMed] [Google Scholar]
- King TH, Decatur WA, Bertrand E, Maxwell ES, Fournier MJ. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol Cell Biol. 2001;21(22):7731–7746. doi: 10.1128/MCB.21.22.7731-7746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner J, Vissi E, Gross S, Szoor B, Rudenko A, Alphey L, White-Cooper H. Drosophila Uri, a PP1alpha binding protein, is essential for viability, maintenance of DNA integrity and normal transcriptional activity. BMC Mol Biol. 2008;9(1):36. doi: 10.1186/1471-2199-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30(5):235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9(2):148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Liu L, McKeehan WL. Sequence analysis of LRPPRC and its SEC1 domain interaction partners suggests roles in cytoskeletal organization, vesicular trafficking, nucleocytosolic shuttling, and chromosome activity. Genomics. 2002;79(1):124–136. doi: 10.1006/geno.2001.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin VF, Stirling PC, Gomez-Reino J, Mwenifumbo JC, Obst JM, Valpuesta JM, Leroux MR. Molecular clamp mechanism of substrate binding by hydrophobic coiled-coil residues of the archaeal chaperone prefoldin. Proc Natl Acad Sci USA. 2004;101(13):4367–4372. doi: 10.1073/pnas.0306276101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Huang Y, Sheikh MS. Cloning and characterization of a novel gene PDRG that is differentially regulated by p53 and ultraviolet radiation. Oncogene. 2003;22(46):7247–7257. doi: 10.1038/sj.onc.1207010. [DOI] [PubMed] [Google Scholar]
- Markus SM, Taneja SS, Logan SK, Li W, Ha S, Hittelman AB, et al. Identification and characterization of ART-27, a novel coactivator for the androgen receptor N terminus. Mol Biol Cell. 2002;13(2):670–682. doi: 10.1091/mbc.01-10-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr MT, 2nd, Isogai Y, Wright KJ, Tjian R. Coactivator cross-talk specifies transcriptional output. Genes Dev. 2006;20(11):1458–1469. doi: 10.1101/gad.1418806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGilvray R, Walker M, Bartholomew C. UXT interacts with the transcriptional repressor protein EVI1 and suppresses cell transformation. FEBS J. 2007;274(15):3960–3971. doi: 10.1111/j.1742-4658.2007.05928.x. [DOI] [PubMed] [Google Scholar]
- McKeegan KS, Debieux CM, Boulon S, Bertrand E, Watkins NJ. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol Cell Biol. 2007;27(19):6782–6793. doi: 10.1128/MCB.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao T, Woychik NA. RNA polymerase subunit RPB5 plays a role in transcriptional activation. Proc Natl Acad Sci USA. 1998;95(26):15281–15286. doi: 10.1073/pnas.95.26.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, et al. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell. 2009;34(2):168–178. doi: 10.1016/j.molcel.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss TN, Vo A, McKeehan WL, Liu L. UXT (Ubiquitously Expressed Transcript) causes mitochondrial aggregation. In Vitro Cell Dev Biol Anim. 2007;43(3–4):139–146. doi: 10.1007/s11626-007-9016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DR, Kuhn JF, Shanab GM, Maxwell ES. Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA. 2000;6(6):861–879. doi: 10.1017/S1355838200992446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Saeki M, Xu L, Nakahara H, Saijo M, Tanaka K, Kamisaki Y. RPAP3 interacts with Reptin to regulate UV-induced phosphorylation of H2AX and DNA damage. J Cell Biochem. 2009;106(5):920–928. doi: 10.1002/jcb.22073. [DOI] [PubMed] [Google Scholar]
- Nikolov DB, Burley SK. RNA polymerase II transcription initiation: a structural view. Proc Natl Acad Sci USA. 1997;94(1):15–22. doi: 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwachukwu JC, Li W, Pineda-Torra I, Huang HY, Ruoff R, Shapiro E, et al. Transcriptional regulation of the androgen receptor cofactor androgen receptor trapped clone-27. Mol Endocrinol. 2007;21(12):2864–2876. doi: 10.1210/me.2007-0094. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000;407(6803):471–476. doi: 10.1038/35035000. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10(21):2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Parsons GG, Spencer CA. Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol Cell Biol. 1997;17(10):5791–5802. doi: 10.1128/mcb.17.10.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parusel CT, Kritikou EA, Hengartner MO, Krek W, Gotta M. URI-1 is required for DNA stability in C. elegans. Development. 2006;133(4):621–629. doi: 10.1242/dev.02235. [DOI] [PubMed] [Google Scholar]
- Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22(2):153–183. doi: 10.1210/er.22.2.153. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386(6625):569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Puri T, Wendler P, Sigala B, Saibil H, Tsaneva IR. Dodecameric structure and ATPase activity of the human TIP48/TIP49 complex. J Mol Biol. 2007;366(1):179–192. doi: 10.1016/j.jmb.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Lin YL, Thome KC, Pian P, Schlegel BP, Weremowicz S, et al. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J Biol Chem. 1998;273(43):27786–27793. doi: 10.1074/jbc.273.43.27786. [DOI] [PubMed] [Google Scholar]
- Roch F, Serras F, Cifuentes FJ, Corominas M, Alsina B, Amorós M, et al. Screening of larval/pupal P-element induced lethals on the second chromosome in Drosophila melanogaster: clonal analysis and morphology of imaginal discs. Mol Gen Genet. 1998;257(2):103–112. doi: 10.1007/PL00008620. [DOI] [PubMed] [Google Scholar]
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579(4):909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Saeki M, Irie Y, Ni L, Yoshida M, Itsuki Y, Kamisaki Y. Monad, a WD40 repeat protein, promotes apoptosis induced by TNF-alpha. Biochem Biophys Res Commun. 2006;342(2):568–572. doi: 10.1016/j.bbrc.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Samanta AK, Huang HJ, Bast RC, Jr, Liao WS. Overexpression of MEKK3 confers resistance to apoptosis through activation of NFkappaB. J Biol Chem. 2004;279(9):7576–7583. doi: 10.1074/jbc.M311659200. [DOI] [PubMed] [Google Scholar]
- Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci USA. 2008;105(5):1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, et al. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101(2):199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Schmidt C, Peng B, Li Z, Sclabas GM, Fujioka S, Niu J, et al. Mechanisms of proinflammatory cytokine-induced bi-phasic NF-kappaB activation. Mol Cell. 2003;12(5):1287–1300. doi: 10.1016/S1097-2765(03)00390-3. [DOI] [PubMed] [Google Scholar]
- Schroer A, Schneider S, Ropers H, Nothwang H. Cloning and characterization of UXT, a novel gene in human Xp11, which is widely and abundantly expressed in tumor tissue. Genomics. 1999;56(3):340–343. doi: 10.1006/geno.1998.5712. [DOI] [PubMed] [Google Scholar]
- Siegers K, Waldmann T, Leroux MR, Grein K, Shevchenko A, Schiebel E, Hartl FU. Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin-GimC system. EMBO J. 1999;18(1):75–84. doi: 10.1093/emboj/18.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegers K, Bölter B, Schwarz JP, Böttcher UM, Guha S, Hartl FU. TRiC/CCT cooperates with different upstream chaperones in the folding of distinct protein classes. EMBO J. 2003;22(19):5230–5240. doi: 10.1093/emboj/cdg483. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Siegert R, Leroux MR, Scheufler C, Hartl FU, Moarefi I. Structure of the molecular chaperone prefoldin: unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell. 2000;103(4):621–632. doi: 10.1016/S0092-8674(00)00165-3. [DOI] [PubMed] [Google Scholar]
- Sun S, Tang Y, Lou X, Zhu L, Yang K, Zhang B, et al. UXT is a novel and essential cofactor in the NF-κB transcriptional enhanceosome. J Cell Biol. 2007;178(2):231–244. doi: 10.1083/jcb.200611081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja SS, Ha S, Swenson NK, Torra IP, Rome S, Walden PD, et al. ART-27, an androgen receptor coactivator regulated in prostate development and cancer. J Biol Chem. 2004;279(14):13944–13952. doi: 10.1074/jbc.M306576200. [DOI] [PubMed] [Google Scholar]
- Te J, Jia L, Rogers J, Miller A, Hartson SD. Novel subunits of the mammalian Hsp90 signal transduction chaperone. J Proteome Res. 2007;6(5):1963–1973. doi: 10.1021/pr060595i. [DOI] [PubMed] [Google Scholar]
- Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77(1):5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Triezenberg SJ. Structure and function of transcriptional activation domains. Curr Opin Genet Dev. 1995;5(2):190–196. doi: 10.1016/0959-437X(95)80007-7. [DOI] [PubMed] [Google Scholar]
- Uhlik MT, Abell AN, Johnson NL, Sun W, Cuevas BD, Lobel-Rice KE, et al. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat Cell Biol. 2003;5(12):1104–1110. doi: 10.1038/ncb1071. [DOI] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93(5):863–873. doi: 10.1016/S0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132(6):945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JL, Etemad-Moghadam B, Guo S, Boyd L, Draper BW, Mello CC, et al. par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development. 1996;122(10):3133–3140. doi: 10.1242/dev.122.10.3133. [DOI] [PubMed] [Google Scholar]
- Wei W, Gu JX, Zhu CQ, Sun FY, Dorjsuren D, Lin Y, Murakami S. Interaction with general transcription factor IIF (TFIIF) is required for the suppression of activated transcription by RPB5-mediating protein (RMP) Cell Res. 2003;13(2):111–120. doi: 10.1038/sj.cr.7290155. [DOI] [PubMed] [Google Scholar]
- Yang J, Lin Y, Guo Z, Cheng J, Huang J, Deng L, et al. The essential role of MEKK3 in TNF-induced NF-kappaB activation. Nat Immunol. 2001;2(7):620–624. doi: 10.1038/89769. [DOI] [PubMed] [Google Scholar]
- Zariwala M, O’Neal WK, Noone PG, Leigh MW, Knowles MR, Ostrowski LE. Investigation of the possible role of a novel gene, DPCD, in primary ciliary dyskinesia. Am J Respir Cell Mol Biol. 2004;30(4):428–434. doi: 10.1165/rcmb.2003-0338RC. [DOI] [PubMed] [Google Scholar]
- Zhao H, Wang Q, Zhang H, Liu Q, Du X, Richter M, Greene MI. UXT is a novel centrosomal protein essential for cell viability. Mol Biol Cell. 2005a;16(12):5857–5865. doi: 10.1091/mbc.E05-08-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005b;120(5):715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Zhao R, Kakihara Y, Gribun A, Huen J, Yang G, Khanna M, et al. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J Cell Biol. 2008;180(3):563–578. doi: 10.1083/jcb.200709061. [DOI] [PMC free article] [PubMed] [Google Scholar]