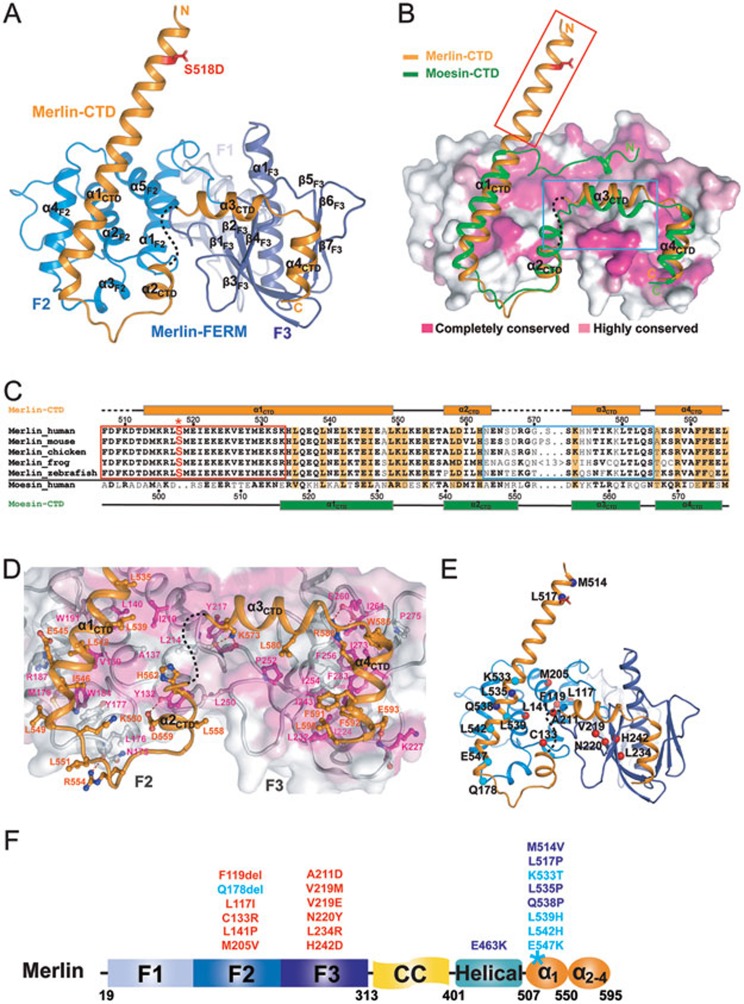
Figure 2.
The overall structure of the Merlin-FERM/Merlin-CTD complex. (A) Ribbon diagram showing the crystal structure of the stabilized, fully closed Merlin-FERM/Merlin-CTD complex. S518D is shown in the stick model and colored in red. (B) Comparison of the structures of the Merlin FERM/CTD complex and the Moesin FERM/CTD complex (PDB code: 1EF1). In this representation, the Merlin-CTD and Moesin-CTD are shown as ribbons, and the Merlin-FERM is drawn in the surface model based on the degree of their amino acid sequence conservation between Merlin and ERMs. The N-terminal half of Merlin α1CTD is boxed in red to emphasize its structural uniqueness. (C) Sequence alignment of the CTD regions from Merlin across different species and human Moesin-CTD. Secondary structural elements for Merlin-CTD and Moesin-CTD are indicated above and below the alignment, respectively. Residues involved in the auto-inhibition of Merlin are emphasized in orange and S518 is highlighted in red and with an asterisk. The residues within the red box form the first half of α1CTD highlighted in red box in B. (D) Molecular details of the Merlin FERM/CTD interaction. (E, F) Ribbon-dot model (E) and a schematic diagram (F) summarizing 21 human cancer-related missense mutations. The mutations that are predicted to interfere with the folding of FERM are shown in red; the mutations that may alter the auto-inhibition and/or AMOT binding of Merlin are colored in cyan and blue.