FIGURE 1.
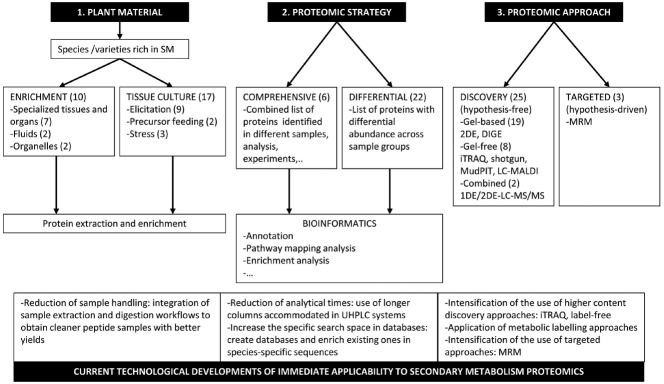
Major issues in the proteomic analysis of plant secondary metabolism. Three major issues have been considered in the proteomic analysis of plant secondary metabolism. Two are common to any type of proteomic analysis, i.e., the strategy to find proteins of interest, and the technical approach to achieve it. In the scheme above, we have included the ways in which such issues have been resolved to date and the corresponding number of representative studies (figure in brackets). So the first and specific issue for accessing the plant secondary metabolism proteome is the selection of suitable plant material, which is rich in secondary metabolites of interest. If using whole plants as a source, collection of specialised tissues-roots, fruit exocarp and mesocarp-, organs –trichomes-, fluids –milky sap- or preparation of organelles –chromoplasts- before starting protein extraction has been a successful strategy to access the target proteome. Alternatively, in vitro cell culture has been a smart option to easily generate an abundant population of homogeneous cells that produce SM whenever they were stimulated through different treatments, such as elicitation, precursor feeding or physical stress. A second issue is the proteomic strategy to find target proteins; i.e., enzymes and transporters specifically involved in the metabolic pathway of interest. One is a comprehensive analysis in which the identification of the largest possible number of proteins is intended. The other typical strategy is differential proteomics. In this case, the proteome complements obtained from two experimental groups or more, which differ in secondary metabolite content, are compared. Proteins with differential abundance are selected. In both cases, a bioinformatics-based analysis of the protein lists follows to classify proteins according to their molecular and (potential) biological function, and to select the candidate proteins involved in SM for further functional characterization using biochemical and genetic tools. Eventually, a third major issue is the proteomic approach. As the initial goal is to find the new enzymes and transporters involved in secondary metabolite synthesis and biology, a hypothesis-free type discovery proteomics approach, either top-down or bottom-up, is usually undertaken. A number of applications of classical and advanced gel-based and gel-free proteomic techniques to investigate plant SM pathways have been reported. Having identified the proteins of interest, a hypothesis-driven targeted proteomics approach is the next step to profoundly characterize the pathway under different experimental conditions. For this purpose, proteomic workflows have utilized MRM. Indeed a number of technological developments of immediate applicability that are currently used in proteomics from which SM proteomic research would very much benefit are suggested. These may introduce advantages in handling plant material to obtain cleaner and higher yield protein or peptide samples, and to provide improvements in analytical times, protein identification rates, and quantification of protein changes at either the whole or targeted proteome level.
