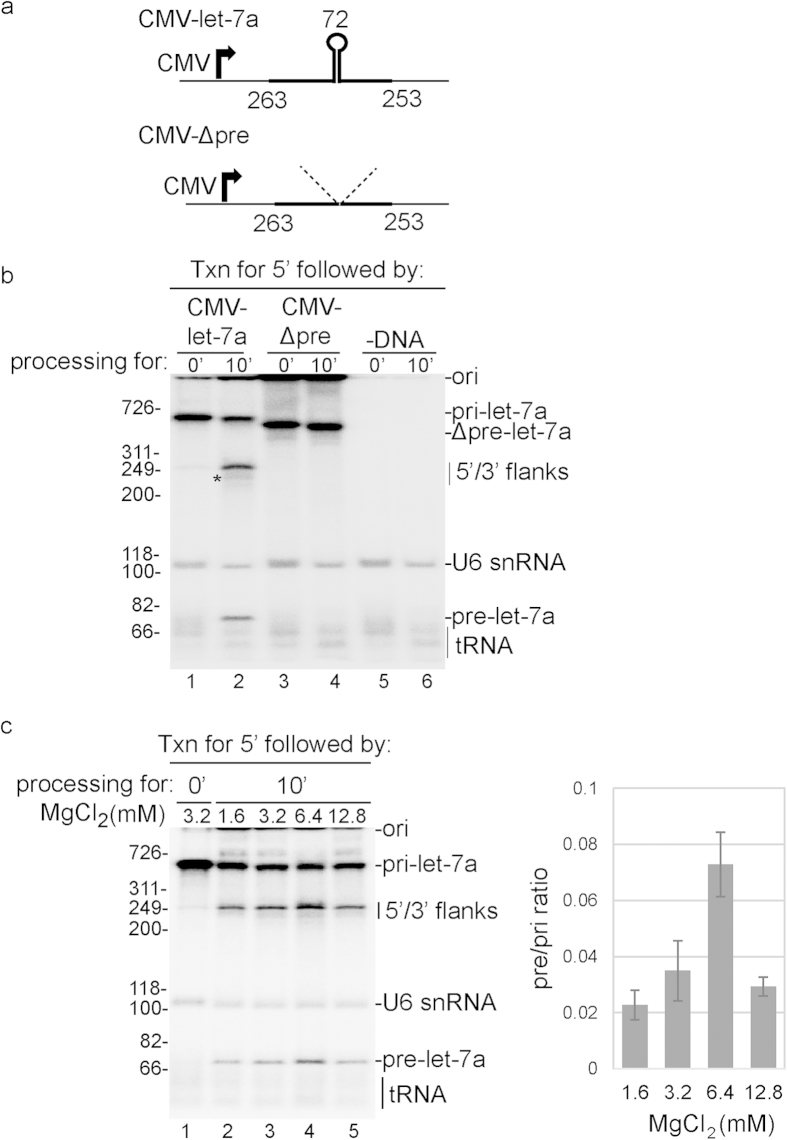
Figure 1. RNAP II txn/pri-miRNA processing system in vitro.
(a) Structure of the CMV DNA template encoding let-7a pri-miRNA used for txn/pri-miRNA processing. The sizes of 5′ flanking region, 3’ flanking region, and pre-let-7a are indicated. The thick line indicates the natural pri-miRNA sequences and the thin lines indicate the vector sequences. CMV-Δpre-let-7a is a mutant lacking the 72 nt pre-let-7a hairpin. (b) RNAP II txn/pri-miRNA processing was carried out for the indicated times using the CMV DNA template encoding let-7a pri-miRNA (lanes 1, 2), the CMV DNA template encoding let-7a Δpre-miRNA DNA template (lanes 3, 4) or no DNA template (lanes 5, 6). The asterisk indicates the 3′ flanking region, which lacks a cap and is therefore largely degraded during the reaction (lane 2). (c) RNAP II txn/pri-miRNA processing was carried using different amounts of MgCl2 as indicated and the CMV DNA template encoding let-7a pri-miRNA. Size markers (in base pairs) and RNA species are indicated. Ori indicates the gel origin. RNAs were detected by phosphorimager and quantified using Quantity One software. Mean ± S.D. of three biological replicates are shown.