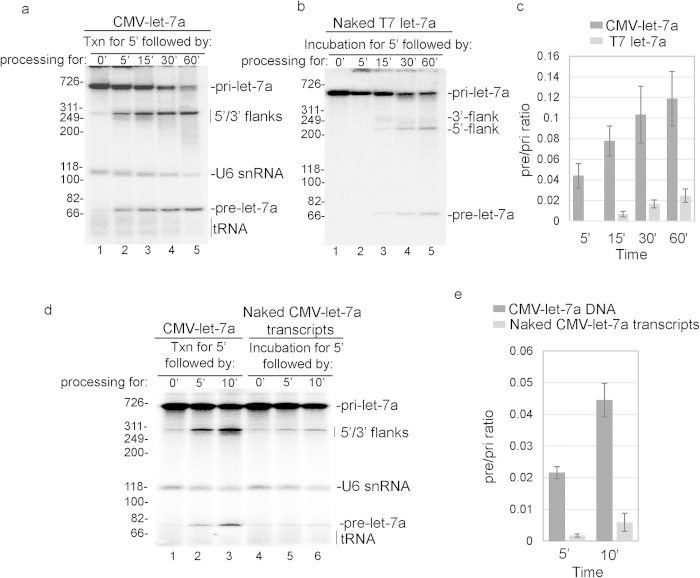
Figure 3. Let-7a pri-miRNA processing is functionally coupled to RNAP II transcription.
(a,b) CMV DNA template encoding let-7a pri-miRNA (a) or the corresponding T7 let-7a pri-miRNA (b) was incubated in nuclear extract in the txn/pri-miRNA processing system for the indicated times. Markers (in base pairs) and RNA species are indicated. (c) RNAs were detected by phosphorimager and quantified using Quantity one software. The ratios of pre-let-7a signal at each point (lanes 2–5) to pri-let-7a signal at the start of the reaction (lane 1) were calculated and mean ± S.D. of three biological replicates are shown. The difference in pre/pri ratio between the two groups at each time point was analyzed by Student’s t-test.(d) CMV DNA template encoding let-7a pri-miRNA (lanes 1–4) or naked CMV let-7a pri-miRNA (lanes 5–8) was incubated in nuclear extract under txn/pri-miRNA reaction conditions for the indicated times. Markers (in base pairs) and RNA species are indicated. Ori marks the gel origin. (e) Same as c except that the data in panel d were quantitated. Mean ± S.D. of three biological replicates are shown, and the difference in pre/pri ratio between the two groups was examined using Student’s t-test.