Table 3. List of 25 DE miRNAs in LD muscle between Yorkshire pigs with low and high RFI.
| Ssc miRNA | Ref miRNA | FC(L/H) | p-value | q-value | Mature Sequence |
|---|---|---|---|---|---|
| ssc-miR-215-5p | hsa-miR-215-5p | −673.96 | 4.57E-27 | 1.48E-24 | augaccuaugaauugacagaca |
| ssc-new-1 | hsa-miR-141-3p | −23.34 | 2.12E-04 | 1.73E-02 | uaacacugucugguaaagaug |
| ssc-miR-194-5p | hsa-miR-194-5p | −19.66 | 8.87E-09 | 1.44E-06 | uguaacagcgacuccauguggac |
| ssc-miR-365-3p | hsa-miR-365a-3p | −1.94 | 0.09 | 1 | uaaugccccuaaaaauccuuau |
| ssc-new-2 | hsa-miR-1249 | −1.74 | 0.41 | 1 | acgcccuucccccccuucuuca |
| ssc-miR-486-5p | hsa-miR-486-5p | −1.69 | 0.04 | 1 | uccuguacugagcugccccgag |
| ssc-miR-1271-5p | hsa-miR-1271-5p | −1.64 | 0.13 | 1 | cuuggcaccuaguaagcacuca |
| ssc-miR-145-5p | hsa-miR-145-5p | −1.6 | 0.18 | 1 | guccaguuuucccaggaaucccu |
| ssc-miR-99b-5p | hsa-miR-99b-5p | −1.51 | 0.17 | 1 | cacccguagaaccgaccuugcg |
| ssc-miR-191-5p | hsa-miR-191-5p | −1.5 | 0.15 | 1 | caacggaaucccaaaagcagcug |
| ssc-miR-10b-5p | hsa-miR-10b-5p | −1.45 | 0.14 | 1 | uacccuguagaaccgaauuugu |
| ssc-miR-29c-3p | hsa-miR-29c-3p | 1.48 | 0.35 | 1 | uagcaccauuugaaaucgguua |
| ssc-miR-92b-3p | hsa-miR-92b-3p | 1.5 | 0.44 | 1 | uauugcacucgucccggccucc |
| ssc-miR-101-3p | hsa-miR-101-3p | 1.55 | 0.19 | 1 | uacaguacugugauaacugaag |
| ssc-miR-338-3p | hsa-miR-338-3p | 1.56 | 0.37 | 1 | uccagcaucagugauuuuguu |
| ssc-miR-199b-3p | hsa-miR-199b-3p | 1.61 | 0.15 | 1 | acaguagucugcacauugguu |
| ssc-new-3 | hsa-miR-130a-3p | 1.73 | 0.25 | 1 | cagugcaauaguauugucaaagc |
| ssc-miR-208b-3p | hsa-miR-208b-3p | 1.74 | 0.11 | 1 | auaagacgaacaaaagguuugu |
| ssc-miR-1-3p | hsa-miR-1 | 1.81 | 0.09 | 1 | uggaauguaaagaaguauguau |
| ssc-miR-30e-5p | hsa-miR-30e-5p | 1.83 | 0.04 | 1 | uguaaacauccuugacuggaagcu |
| ssc-miR-335-3p | hsa-miR-335-3p | 1.89 | 0.28 | 1 | uuuuucauuauugcuccugacc |
| ssc-miR-136-3p | hsa-miR-136-3p | 1.93 | 0.12 | 1 | caucaucgucucaaaugagucu |
| ssc-new-4 | hsa-miR-144-3p | 2.1 | 0.32 | 1 | uacaguauagaugauguacu |
| ssc-new-5 | hsa-miR-301b | 2.15 | 0.46 | 1 | cagugcaaugauauugucaaagc |
| ssc-new-6 | hsa-miR-190a | 2.28 | 0.34 | 1 | ugauauguuugauauauuagguug |
The p-value was calculated by the formula: 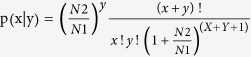 with a Python script.
with a Python script.
The q-value was calculated by “fdrtool” in R, and FDR (false discovery rate) ≤ 0.05 for DEG significant determination.
