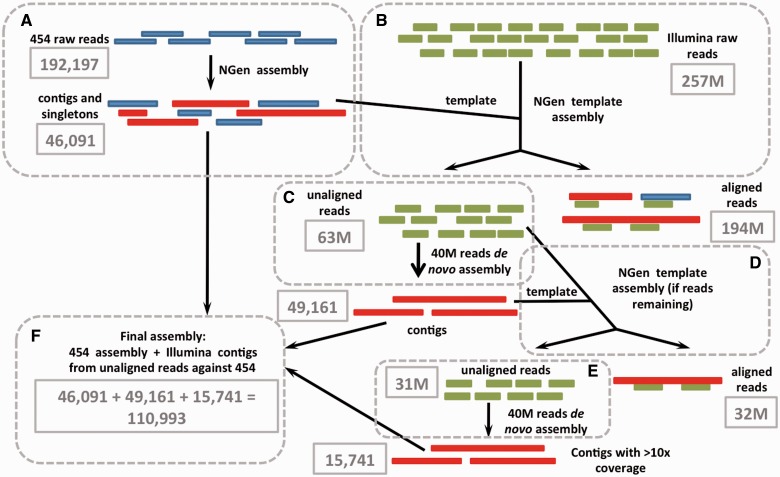
Fig. 2.—
Assembly pipeline used to combine the “454” (in blue) and Illumina (in green) reads into nonredundant contigs (in red). Framed values correspond to those obtained for the E. macularius mix data set (multiple developmental stages and multiple adult organs), provided as an example. Dashed boxes delineate major steps of the assembly: (A) The “454” reads are de novo assembled, (B) the Illumina reads are aligned to the “454” assembly, and (C–E) iterative building of nonredundant Illumina contigs. The final assembly (F) includes both the “454” assembly and the Illumina contigs.