Fig. 1.—
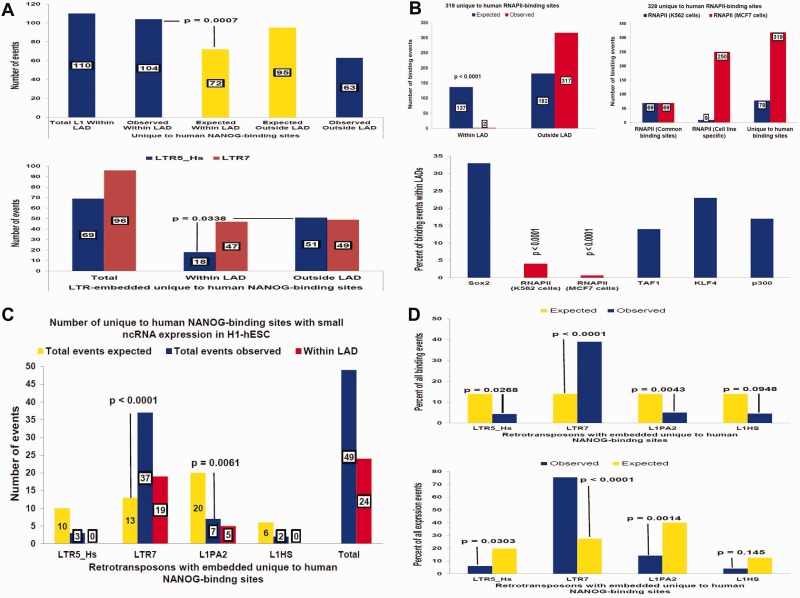
Characterization of genomic features associated with human ESC-specific NANOG- and CTCF-binding sites. Location of full-length L1 TE sequences containing human ESC-specific NANOG-binding sites is enriched within LADs (A). In the hESC genome, there are 184 human-specific NANOG-binding sites that are embedded within 167 full-length and 17 truncated L1HS and L1PA2 sequences, 110 of which are located within LADs (A, top panel). In total, 104 of 110 (95%) of the L1HS and L1PA2 sequences containing human-specific NANOG-binding sites and located within LADs are preserved as full-length (5,962–6,189 bp) L1 retrotransposons (A, top panel), indicating that the conservation of the full-length L1 TE containing human-specific NANOG-binding sites is significantly higher within LADs. In contrast, the majority of LTR5_HS-embedded human-specific NANOG-binding sites is found outside of LADs, whereas LTR7-embedded human-specific NANOG-binding sites are equally distributed within and outside of LADs (A, bottom panel). Distinct placement patterns within LADs of 446 human-specific TF-binding sites for five different regulatory proteins (B). Note that a significantly higher fraction than expected by chance of RNAPII-binding sites is located outside of LADs (B) in contrast to other TF-binding sites. LTR7-derived sncRNAs represent the predominant type of sncRNAs generated by transcriptional activity of the human-specific NANOG-binding sites in hESCs and are transcribed from the loci equally distributed within and outside the LADs (C, D).