Fig. 3.—
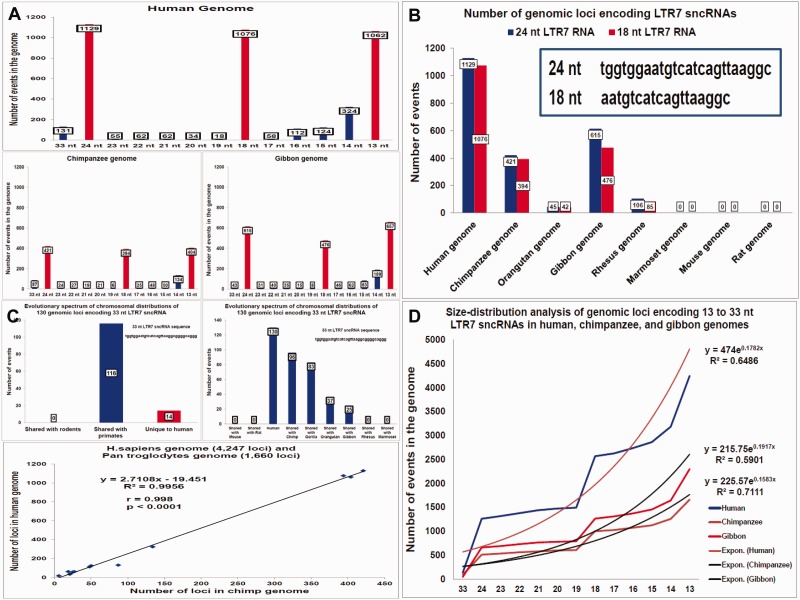
Analysis of LTR7-derived sncRNAs transcribed from human-specific regulatory loci reveals evolutionary conservation of wild-type sequences. Distinct molecular variants of the 33- and 24-nt LTR7 sncRNAs manifest a primate-specific evolutionary spectrum of chromosomal distributions (A–D) and highly correlated patterns of chromosomal distributions in the human genome (r = 0.865; P < 0.0001; supplementary fig. S4, Supplementary Material online). Size distribution analyses in the human and primate genomes of fully conserved LTR7 sncRNA-encoding loci of various lengths revealed essentially identical profiles of genome-wide size distributions of the LTR7 sncRNA-encoding loci in the human, chimpanzee, and gibbon genomes (A) with identical 24- and 18-nt sequences representing the predominantly conserved molecular entities (A, B), which are significantly overrepresented in the human genome compared with the primate and rodent genomes (A, B, D). Panel (A) shows results of the size distribution analysis and panel (D) shows the linear regression analysis in the rodent, human, and primate genomes of fully conserved LTR7 sncRNA-encoding loci of differing lengths. Note the essentially identical profiles of genome-wide size distributions of the LTR7 sncRNA-encoding loci in the human, chimpanzee, and gibbon genomes (C, bottom panel; r = 0.998; P < 0.0001), with identical 24- and 18-nt sequences representing the predominantly conserved molecular entities (A, D), which are significantly overrepresented in the human genome compared with the primate and rodent genomes. Panel (C) (top two panels) shows evolutionary conservation in primates of chromosomal positions of the wild-type 33-nt LTR7 sncRNA-encoding sequences.