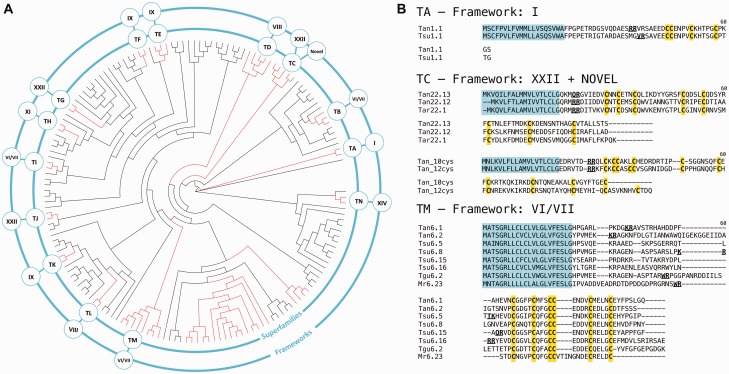
Fig. 6.—
Teretoxin gene superfamilies. (A) Circular phylogeny of terebrid gene superfamilies (midpoint root) obtained from MUSCLE alignment of teretoxin signal sequences under ML criteria with JTT + G and 1,000 pseudoreplicates. Clades highlighted in red have strong support values but only those that meet additional criteria (see details in text) are designated as teretoxin superfamilies. Inner ring shows superfamily name and outer ring Cys framework of the superfamily. (B) Sequence alignment of representative teretoxin superfamilies and their associated frameworks. Note that superfamily TC includes teretoxins with Cys framework XXII (top) and two novel frameworks (bottom). Also, superfamily TM shares significant sequence identity with the H superfamily of conotoxins (included as an example Mr6.23 from C. marmoreus) Tan, Tr. anilis; Tsu, Te. subulata; Tar, Te. argosyia; Tgu, Te. guttata.