Fig. 4.
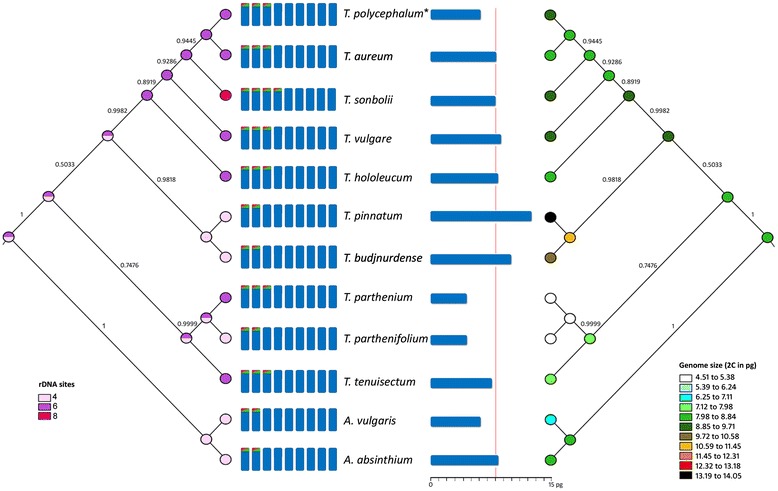
Ancestral state reconstruction of number of rDNA signals (left) and genome size (right) for diploid Tanacetum taxa. The model of reconstruction was Parsimony as implemented in Mesquite (v.3.02), and ancestral state reconstruction was estimated using the 50 % majority-rule consensus topology obtained by Bayesian inference phylogenetic analysis of the internal transcribed spacer 1 (ITS1), ITS2 and trnH-psbA data sequence. The Bayesian clade-credibility values (posterior probability > 0.5) are given above branches. Schematic representation of chromosomes with the most commonly found numbers of rDNA signals and bars that depict genome sizes (2C values) with a red line indicating the mean 2C value at the diploid level. (*) Tanacetum polycephalum ssp. argyrophyllum
