Abstract
High resolution X-ray crystal structures of multisubunit RNA polymerases (RNAP) have contributed to our understanding of transcriptional mechanisms. They also provided a powerful guide for the design of experiments aimed at further characterizing the molecular stages of the transcription reaction. Our laboratory used tandem-affinity peptide purification in native conditions to isolate human RNAP II variants that had site-specific mutations in structural elements located strategically within the enzyme’s catalytic center. Both in vitro and in vivo analyses of these mutants revealed novel features of the catalytic mechanisms involving this enzyme.
Keywords: RNA polymerase II, transcriptional mechanisms, mutational analysis, mRNA synthesis
Introduction
In prokaryotes, a single RNA polymerase (RNAP) synthesizes all classes of RNA, including the messenger RNA (mRNA), the ribosomal RNA (rRNA), and the transfer RNA (tRNA). Prokaryotic RNAPs are composed of 5 subunits (α2, β′, β, and ω), which form the core enzyme; the core enzyme associates with a σ factor to form the holoenzyme required for promoter recognition (Burgess and Travers 1970). In eukaryotes, RNAP I synthesizes the rRNA, RNAP II forms the mRNA and the small nuclear RNA, and RNAP III is responsible for the synthesis of the tRNA and the 5s rRNA. RNAP I, II, and III contain 14, 12, and 17 subunits, respectively, of which 4 are related and 5 are identical (Carles et al. 1991; Woychik and Young 1990). In each case, the 2 largest subunits (bacterial RNAP β′ and β, RNAP I Rpa190 and Rpa135, RNAP II Rpb1 and Rpb2, RNAP III Rpc160 and Rpc128) carry the catalytic activity of the enzyme and share sequence homologies. The multisubunit archaebacterial RNAP is composed of 13 polypeptides, the 3 largest subunits of which are the homologues of the 2 largest subunits of the eukaryotic and prokaryotic RNAPs (Langer et al. 1995). Six other archaeal subunits share sequence homologies with polypeptides of bacterial and eukaryotic RNAPs.
The resolution of the crystallographic structures of multi-subunit RNAPs has helped to elucidate transcriptional mechanisms. To date, high-resolution structures are available for the bacterial RNAP and the eukaryotic RNAP II alone (Cramer et al. 2001; Armache et al. 2003; Bushnell and Kornberg 2003; Zhang et al. 1999), and as part of a complex with nucleic acids (Westover et al. 2004a, 2004b; Gnatt et al. 2001; Murakami et al. 2002a; Kettenberger et al. 2004) or regulatory factors (Kettenberger et al. 2003; Bushnell et al. 2004; Opalka et al. 2003; Vassylyev et al. 2002; Murakami et al. 2002b). Both the prokaryotic and the eukaryotic transcription reactions involve a number of steps, starting with promoter recognition. The bacterial σ factor (Gruber and Gross 2003) and the eukaryotic general transcription factors (GTF) TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (Hampsey 1998; Coulombe and Burton 1999) are required for specific binding of the polymerase to the promoter of a gene. In both systems, promoter binding is accompanied by bending and wrapping of the promoter DNA against the body of the polymerase (Forget et al. 1997; Langelier et al. 2001; Robert et al. 1998; Rivetti et al. 1999, 2003). Our laboratory has shown that DNA wrapping is involved in promoter melting in the region of the transcriptional initiation site, probably through the induction of a torsional strain that leads to the unwinding of the DNA double helix (Douziech et al. 2000; Forget et al. 2004). The formation of a left-handed loop in the promoter DNA wrapped around the mobile clamp of the enzyme, as revealed by our recent site-specific protein–DNA photo-cross-linking experiments (Forget et al. 2004), was proposed to induce DNA unwinding, which is then stabilized by some GTFs. The formation of this open complex allows the pairing of incoming ribonucleoside triphosphates to the template DNA strand for phosphodiester bond formation (Holstege et al. 1996, 1997; Craig et al. 1998). RNAP then starts a cycle of abortive initiation, where it synthesizes and releases small transcripts without disengaging from the DNA template (Vo et al. 2003). When the transcript reaches a length of approximately 10 to 12 nucleotides, the transcription complex is stabilized and RNAP breaks its contacts with the GTF/σ and clears the promoter for elongation (Holstege et al. 1997; Craig et al. 1998).
Structure of multisubunit RNA polymerases
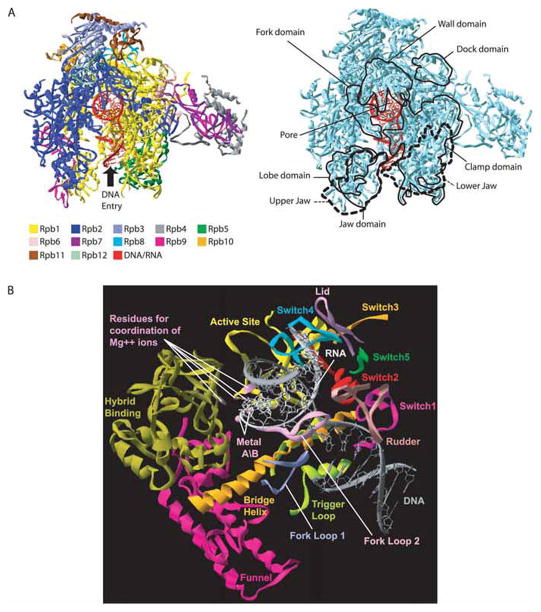
Crystallographic studies have revealed that the structure of RNAP is highly conserved from prokaryotes to eukaryotes, with the regions of highest homology mapping near the active center (Cramer et al. 2001; Zhang et al. 1999). The catalytic subunits (β′, β; Rpb1, Rpb2) constitute the body of the polymerase, in which a positively charged cleft is centered (Cramer et al. 2001; Zhang et al. 1999). The 2 Mg2+ ions, metals A and B, responsible for the catalysis are buried deep in this cleft, which accommodates the nucleic acids during transcription (Fig. 1A) (Westover et al. 2004b). Steitz (1998) proposed a 2-metal ion mechanism underlying the catalytic activity of all the polymerases. A domain of Rpb2/β, called the wall, closes the upstream extremity of this primary channel. The nucleic acids are retained in the cleft by the clamp domain and by a number of protein domains, including the upper and lower jaws, which grab the downstream DNA (Gnatt et al. 2001). Different crystallographic structures have indicated that promoter DNA is first loaded on the top of the clamp (Armache et al. 2003; Bushnell and Kornberg 2003; Murakami et al. 2002a) The template strand reaches the active site only after promoter melting has occurred. A secondary channel, called the pore, has the shape of an inverted funnel, which opens in the cleft near the active site. It has been proposed that the incoming nucleoside triphosphate (NTP) and the released pyrophosphate circulate through this pore (Cramer et al. 2001; Gnatt et al. 2001). The mRNA exit channel lies between the wall and the clamp domains.
Fig. 1.
High-resolution X-ray crystal structure of eukaryotic RNA polymerase (RNAP) II. (A) Structure of Saccharomyces cerevisiae RNAP II in complex with nucleic acids (PDB accession number 1Y1W). Left panel: The various subunits and the nucleic acids are shown (see color code). The DNA enters the RNAP II structure through the cleft as indicated, turns by about 90 degrees, and exit perpendicular to the plane. Right panel: The main structural domains of the enzyme are indicated. (B) Structural elements surrounding the DNA–RNA hybrid in RNAP II. The rudder, switch2, funnel, bridge helix, lid, trigger loop, switch1, and switch5 features of Rpb1 are listed. The fork-loop 1, fork-loop 2, switch3, and switch4 features of Rpb2 are indicated.
In addition to their polymerization activity, DNA-dependant RNAPs can also catalyze the 3′ endonucleotydic cleavage of transcripts under certain circumstances (Conaway et al. 2000; Shilatifard et al. 2003). First discovered in Escherichia coli (Krummel and Chamberlin 1989; Surratt et al. 1991), this cleavage activity takes place when RNAP backtracks at pause and arrest sites (Wind and Reines 2000; Fish and Kane 2002). Weakly associated DNA–RNA hybrids can induce backtracking of the enzyme to a more stable register (Nudler et al. 1997). To resume elongation from this more stable position, RNAP needs to cleave the 3′ end of the transcript that extrudes from the catalytic site through the pore. The 3′ cleavage activity is enhanced by cleavage factors GreA and GreB in bacteria (Borukhov et al. 1992, 1993) and TFIIS in eukaryotes (Wind and Reines 2000; Fish and Kane 2002; Reines 1992). The crystal structure of TFIIS in a complex with RNAP II (Kettenberger et al. 2003) has provided insight into the mechanism by which this factor enhances transcript cleavage. More specifically, it revealed that 2 conserved acidic residues in TFIIS, essential for its activity (Jeon et al. 1994), are located in the vicinity of the polymerase metal B, and might participate in its coordination. Therefore, it has been suggested that the 2-metal ion mechanism used for NTP polymerization is also involved in the 3′ endonucleotydic activity of RNAP II. In addition, recent studies on GreB, the E. coli homologue of TFIIS, have provided evidence of a 2-metal ion mechanism in the 3′ transcript cleavage activity of bacterial RNAP (Opalka et al. 2003; Sosunova et al. 2003).
Structures of elongating RNAP have revealed the presence of an 8–9 base-pair DNA–RNA hybrid in the cleft (Westover et al. 2004a; Gnatt et al. 2001). The crystallographic data have also shown that a number of loops and helices of Rpb1/β′ and Rpb2/β are located in this region, in proximity to the catalytic site (Fig. 1B). These structural elements were named according to their location or to their presumed role in the transcriptional process. For example, the Rpb1/β′ bridge helix separates the main channel and is located near the template DNA at the +1 site. Because crystallographic data have revealed 2 different conformations of this structure, bent (Zhang et al. 1999) or straight (Cramer et al. 2001; Westover et al. 2004a; Gnatt et al. 2001), it has been proposed that the bridge helix is involved in the translocation of nucleic acids during transcription. The localization of the rudder, the lid and fork-loop 1 suggests that these loops are involved in DNA–RNA strand separation, by maintaining an 8–9 base-pair hybrid (Westover et al. 2004a). Fork-loop 2 and the zipper might be involved in maintaining of the upstream and downstream ends of the transcription bubble, respectively (Cramer et al. 2001; Gnatt et al. 2001). Five loops of Rpb1 and Rpb2 have been called the switches, and might participate in forming a binding site for the DNA–RNA hybrid and (or) controlling the position of the clamp (Cramer et al. 2001; Armache et al. 2003; Gnatt et al. 2001).
Affinity purification of RNA polymerase II mutants
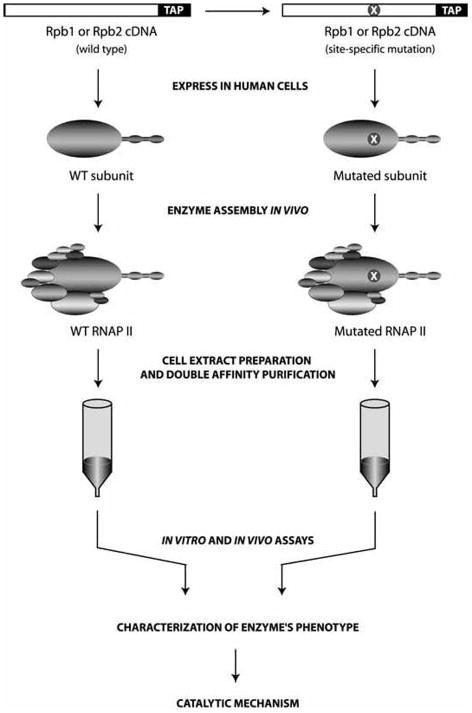
To determine the function of the various elements strategically located within the structure of RNAP II, and to further define their respective roles during the transcription reaction, we programmed human 293 cells to express RNAP II subunits carrying both a specific mutation and a tandem-affinity peptide (TAP) tag at their C-terminus (Jeronimo et al. 2004) (Fig. 2). Following expression of the tagged mutated RNAP II subunit, double-affinity chromatography was used to purify the RNAP II mutant enzyme from cell extracts. Because the tandem-affinity purification procedure is carried out in native conditions, the purified mutants can be analyzed in various functional assays that reconstruct in vitro the various stages of the transcription reaction. Chromatin immunoprecipitation (ChIP) assays were used to analyze the ability of the mutated RNAP II to associate with promoter DNA and to transcribe genes in vivo. In these various assays, each mutated enzyme was compared with the tagged wild-type polymerase, which was also purified using the TAP procedure. Importantly, we showed that wild-type RNAP II, purified using each of the tagged catalytic subunits Rpb1 and Rpb2, is active both in vitro and in vivo. For example, we showed that TAP-tagged RNAP II can assemble into a functional preinitiation complex on promoter DNA to initiate transcription, to escape the promoter, to elongate the transcript, to respond to the cleavage stimulatory factor TFIIS, to assemble on promoter DNA in vivo, and to localize along transcription units in vivo. This extensive characterization of wild-type TAP-tagged RNAP II revealed that the presence of the residual calmodulin-binding peptide tag on RNAP II does not significantly affect the activity of the enzyme.
Fig. 2.
Overview of the procedure for the design and purification of RNAP II mutants. Wild-type (left part) and mutated (right part: X, mutation) RNAP II subunits (Rpb1 or Rpb2) carrying a tandem-affinity peptide (TAP) tag are expressed in human cells, and the corresponding RNAP II complexes are purified using double-affinity chromatography in native conditions. The purified enzymes are submitted to various assays that reconstruct the different steps of the transcription reaction. Because RNAP II variants carry a tag, they can be localized along genomic DNA in vivo using chromatin immunoprecipitation (ChIP) experiments that are followed by PCR detection of enriched regions (promoter or transcribed region). WT, wild type.
Characterization of RNA polymerase II mutants
Guided by the high-resolution structures available for RNAP II (Bushnell and Kornberg 2003; Gnatt et al. 2001; Kettenberger et al. 2003), we used the TAP procedure to purify and characterize many RNAP II mutants and begin a systematic structure–function analysis of this enzyme (Table 1). The mutations studied to date were found to affect various stages of the transcription reaction and are discussed below.
Table 1.
Summary of the characterization of RNAP II mutants.
| RNAPII | In vitro assays
|
In vivo location
|
||||||
|---|---|---|---|---|---|---|---|---|
| RNAPII assembly | PIC formation | Initiation on premelted promoter | Initiation on closed promoter | NTP-Mg(B) binding | Transcript elongation | Promoter | Transcribed region | |
| Rpb2 | ||||||||
| WT | + | + | + | + | + | + | + | + |
| Switch3 (R1078A, S1079A, R1080A) | + | ± | − | − | − | − | − | |
| Fork 1 (Δ458–459) | + | + | ± | − | − | + | − | |
| Conserved E791A (E791A) | + | + | + | + | ± | ± | + | ± |
| Rpb1 | ||||||||
| Rudder (Δ321–334) | + | − | − | − | − | |||
| Zipper (Δ45–55) | + | − | − | − | − | |||
| WT | + | + | + | + | + | |||
Note: Various assays that monitor the different stages of the transcription reaction were used to compare each RNAP II mutant to the wild-type enzyme. Our assays monitored the following: 12-subunit core RNAP II assembly, preinitiation complex (PIC) formation on promoter DNA in the presence of the general transcription factors, abortive initiation on a premelted template (this assay allowed us to bypass the promoter melting requirement), abortive initiation on a closed template, NTP–Mg binding, and elongation using a C-tailed template or a promoter-dependent run off assay. ChIP assays were used to localize the RNAP II variants on the promoter or transcribed region of active genes in vivo. NTP, nucleoside triphosphate; empty boxes, not determined.
The localization of Rpb2 fork-loop 1 suggests a possible role for this element in DNA–RNA strand separation, in such a way that an 8–9 base-pair hybrid is maintained during transcription (Westover et al. 2004a). A fork 1 mutant (fork 1 Δ458–459), lacking 2 residues near the center of the loop, was purified and characterized. This mutant supports the formation of a preinitiation complex with the GTF on the adenovirus major late (AdML) promoter as efficiently as the wild-type enzyme (Jeronimo et al. 2004) (Table 1). However, the fork 1 mutant was unable to initiate transcription from a promoter or to transcribe in a promoter-independent transcription assay that used a C-tailed template. Wild-type RNAP II can initiate transcription from a C-tailed template in the absence of the GTF (Gnatt et al. 1997; Sluder et al. 1988). Because of the assumed role of fork-loop 1 in maintaining the transcription bubble (Cramer et al. 2001; Gnatt et al. 2001), and (or) in DNA–RNA strand separation (Westover et al. 2004a), the fork 1 mutant was tested for its ability to initiate transcription on a template that was artificially melted, between −9 and +2, to mimic the open complex. Interestingly, we observed that artificial melting of the DNA template partly restored the ability of the mutant to initiate transcription. ChIP experiments have shown that the fork 1 mutant can be recruited to the promoter of 2 active genes in vivo, but is absent from the transcribed region of the same genes, supporting our in vitro results (Jeronimo et al. 2004). To explain these results, we proposed that a mutation in fork-loop 1 impairs interactions between the enzyme and the melted DNA at very early stages of the transcription reaction, affecting the ability of the enzyme to initiate transcription efficiently. Given the crystal structure of elongating yeast RNAP II, which shows an interaction between Lys 471 (Lys 458 in human RNAP II) and RNA around positions −5 to −7 (Westover et al. 2004a), an alternative explanation for the defect seen in the fork 1 mutant is that, during transcription initiation, the mobile fork-loop 1 makes crucial contacts with the short elongating RNA, a process essential for phosphodiester bond formation. We speculate that fork-loop 1 stays in contact with the RNA as transcription proceeds, up to the point where it would participate in RNA–DNA strand separation.
Three mutants targeting loops of Rpb1 and Rpb2 — the zipper (zip Δ45–55), the rudder (rud Δ321–334), and switch3 (sw3 R1078A S1079A R1080A) — were found to be defective in the formation of a preinitiation complex on AdML promoter DNA in the presence of the GTFs (Jeronimo et al. 2004; Langelier et al. 2005) (Table 1). Not surprisingly, these 3 mutants were unable to initiate transcription in vitro in any type of assay used. One explanation for the failure of RNAP II mutants in switch3, the zipper, and the rudder to enter a preinitiation complex could be their failure to properly interact with promoter DNA. This would also account for their inability to transcribe from a C-tailed template. Alternatively, it is possible that the mutants interact inaccurately with one of the GTFs involved in the formation of the preinitiation complex. This implies, however, that the mutants have a second defect in elongation, which would explain their inability to transcribe in the GTF-independent C-tailed template assay. Notably, the switch3 loop makes contact with the template DNA at positions −2 and −5, after the template DNA is loaded on the clamp and the promoter is melted and accommodated in the active site cleft (Gnatt et al. 2001; Armache et al. 2004). Because the switch3 mutant is defective in forming the preinitiation complex, our results suggest that switch3 is necessary for the accurate interaction between the clamp and the promoter DNA before the transcription bubble is opened. Consistent with the in vitro data, ChIP experiments have shown that the switch3 mutant is unable to assemble on the promoter of 2 active genes in vivo (Jeronimo et al. 2004).
As mentioned above, multisubunit RNAPs use a 2-metal ion mechanism for polymerization (Steitz 1998). In this mechanism, a first Mg2+ ion, metal A (Mg(A)), lowers the affinity of the 3′ oxygen for hydrogen, thereby facilitating the O− attack on the 5′ α-phosphate. The second Mg2+ ion, metal B (Mg(B)), facilitates the release of the pyrophosphate. In yeast, Mg(A) is coordinated by 3 aspartate residues of Rpb1, D481, D483, and D485, contained within the strictly conserved motif NADFDGD (Cramer et al. 2001; Westover et al. 2004b; Gnatt et al. 2001). Originally, it was proposed that Mg(B) was coordinated by the highly conserved residues E836 and D837 of Rpb2 (Cramer et al. 2001; Gnatt et al. 2001). However, recent structures have localized Mg(B) near residues D481 and D483 of Rpb1, and residue D837 of Rpb2, but too far from the E836 to be coordinated by it (Westover et al. 2004b). A number of studies have also proposed that Mg(B) necessitates the presence of the incoming NTP for the complete coordination required for the polymerization process (Cramer et al. 2001; Westover et al. 2004b; Gnatt et al. 2001; Nedialkov et al. 2003).
To investigate the role of the highly conserved residues E791 and D792 of Rpb2 (equivalent to E836 and D837 in yeast RNAP II), mutants of RNAP II with alanine substitutions in these residues were studied. Mutants MB D792A and MB E791A D792A were unable to assemble in a complete 12-subunit RNAP II enzyme, and therefore were not studied further (not shown). However, mutant MB E791A assembled correctly and was found to be sensitive to both the NTP and Mg2+ concentrations required for NTP polymerization activity, and to Mg2+ concentrations required for the cleavage activity of arrested backtracked transcripts (Langelier et al. 2005).
The structure of RNAP II with a mismatched nucleotide revealed the existence of a second binding site for the incoming NTP, called the entry or E site (Westover et al. 2004b). It has been suggested that the NTP–Mg(B) complex enters the active site through the pore and binds to the E site, first in an inverted orientation, then after rotating, by subsequently binding to the so-called addition or A site. The MB E791A mutant is sensitive to Mg2+ concentrations for polymerization, but the crystallographic data have shown that it is not directly implicated in the coordination of Mg(B) for phosphodiester bond catalysis. Therefore, we proposed a model wherein Rpb2 E791 is implicated in loading the NTP–Mg(B) into the E site (Langelier et al. 2005). This can occur through 2 distinct mechanisms. First, it is possible that the Rpb2 E791 side chain is essential for specifying the topology of the E site; an alanine substitution might modify the configuration of the E site, reducing its binding affinity for the incoming NTP–Mg(B). Second, it is possible that Rpb2 E791 is part of a third, distinct NTP–Mg(B) binding site within the active center, which we called the preloading site (or P site).
The mutant MB E791A is also sensitive to MgCl2 concentration in the cleavage reaction, suggesting that E791 is implicated in binding Mg(B) for this activity. High-resolution structures of RNAP II engaged in transcript cleavage will be required to determine the molecular basis of the interaction between E791 and Mg(B) during the rescue of backtracked transcripts.
Conclusions
Our functional analysis of structural elements located within the catalytic center of RNAP II has allowed us to develop a better understanding of the transcription reaction. In some cases, such as our mutants in the rudder, the zipper, and switch3, a more detailed biochemical characterization will help to define, with precision, their defects (e.g., inaccurate interaction with the DNA or 1 or more GTFs). The TAP-tagging procedure will permit us to characterize a large number of mutants, targeting other structural elements of RNAP II. Notably, the analysis of a series of mutants in the bridge helix, the flap loop, and the trigger loop will most likely reveal novel mechanisms of transcription.
Acknowledgments
We are grateful to the members of our laboratory for helpful discussions. We thank Julie Edwards for critical reading of the manuscript, Diane Bourque for artwork, and Vincent Trinh for the models in Fig. 1. Our laboratory is supported by grants from the Canadian Institutes for Health Research, Genome Canada, and Genome Québec. B.C. is a senior scholar from the Fonds de la recherche en santé du Québec. M.F.L. holds studentships from the Fonds québécois de la recherche sur la nature et les technologies.
Footnotes
This paper is one of a selection of papers published in this Special Issue, entitled 26th International West Coast Chromatin and Chromosome Conference, and has undergone the Journal’s usual peer review process.
References
- Armache KJ, Kettenberger H, Cramer P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc Natl Acad Sci USA. 2003;100:6964–6968. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache KJ, Mitterweger S, Meinhart A, Cramer P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem. 2004;280(8):7131–7134. doi: 10.1074/jbc.M413038200. [DOI] [PubMed] [Google Scholar]
- Borukhov S, Polyakov A, Nikiforov V, Goldfarb A. GreA protein: A Transcription elongation factor from Escherichia coli. Proc Natl Acad Sci USA. 1992;89:8899–8902. doi: 10.1073/pnas.89.19.8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Burgess RR, Travers AA. Escherichia coli RNA polymerase: purification, subunit structure, and factor requirements. Fed Proc. 1970;29:1164–1169. [PubMed] [Google Scholar]
- Bushnell DA, Kornberg RD. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc Natl Acad Sci USA. 2003;100:6969–6973. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science (Wash DC) 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- Carles C, Treich I, Bouet F, Riva M, Sentenac A. Two additional common subunits, ABC10 alpha and ABC10 beta, are shared by yeast RNA polymerases. J Biol Chem. 1991;266:24092–24096. [PubMed] [Google Scholar]
- Conaway JW, Shilatifard A, Dvir A, Conaway RC. Control of elongation by RNA polymerase II. Trends Biochem Sci. 2000;25:375–380. doi: 10.1016/s0968-0004(00)01615-7. [DOI] [PubMed] [Google Scholar]
- Coulombe B, Burton ZF. DNA bending and wrapping around RNA polymerase: a “revolutionary” model describing transcriptional mechanisms. Microbiol Mol Biol Rev. 1999;63:457–478. doi: 10.1128/mmbr.63.2.457-478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig ML, Tsodikov OV, McQuade KL, Schlax PE, Jr, Capp MW, Saecker RM, Record MT., Jr DNA footprints of the two kinetically significant intermediates in formation of an RNA polymerase-promoter open complex: evidence that interactions with start site and downstream DNA induce sequential conformational changes in polymerase and DNA. J Mol Biol. 1998;283:741–756. doi: 10.1006/jmbi.1998.2129. [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 Ångstrom resolution. Science (Wash DC) 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- Douziech M, Coin F, Chipoulet JM, Arai Y, Ohkuma Y, Egly JM, Coulombe B. Mechanism of promoter melting by the xeroderma pigmentosum complementation group B helicase of transcription factor IIH revealed by protein-DNA photo-cross-linking. Mol Cell Biol. 2000;20:8168–8177. doi: 10.1128/mcb.20.21.8168-8177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochim Biophys Acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- Forget D, Robert F, Grondin G, Burton ZF, Greenblatt J, Coulombe B. RAP74 induces promoter contacts by RNA polymerase II upstream and downstream of a DNA bend centered on the TATA box. Proc Natl Acad Sci USA. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget D, Langelier MF, Therien C, Trinh V, Coulombe B. Photo-cross-linking of a purified preinitiation complex reveals central roles for the RNA polymerase II mobile clamp and TFIIE in initiation mechanisms. Mol Cell Biol. 2004;24:1122–1131. doi: 10.1128/MCB.24.3.1122-1131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnatt A, Fu J, Kornberg RD. Formation and crystallization of yeast RNA polymerase II elongation complexes. J Biol Chem. 1997;272:30799–30805. doi: 10.1074/jbc.272.49.30799. [DOI] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science (Wash DC) 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, van der Vliet PC, Timmers HT. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Fiedler U, Timmers HT. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon C, Yoon H, Agarwal K. The transcription factor TFIIS zinc ribbon dipeptide Asp-Glu is critical for stimulation of elongation and RNA cleavage by RNA polymerase II. Proc Natl Acad Sci USA. 1994;91:9106–9110. doi: 10.1073/pnas.91.19.9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Langelier MF, Zeghouf M, Cojocaru M, Bergeron D, Baali D, et al. RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol Cell Biol. 2004;24:7043–7058. doi: 10.1128/MCB.24.16.7043-7058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Krummel B, Chamberlin MJ. RNA chain initiation by Escherichia coli RNA polymerase. Structural transitions of the enzyme in early ternary complexes. Biochemistry. 1989;28:7829–7842. doi: 10.1021/bi00445a045. [DOI] [PubMed] [Google Scholar]
- Langelier MF, Forget D, Rojas A, Porlier Y, Burton ZF, Coulombe B. Structural and functional interactions of transcription factor (TF) IIA with TFIIE and TFIIF in transcription initiation by RNA polymerase II. J Biol Chem. 2001;276:38652–38657. doi: 10.1074/jbc.M106422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Baali D, Trinh V, Greenblatt J, Archambault J, Coulombe B. The highly conserved glutamic acid 791 of Rpb2 is involved in the binding of NTP and Mg(B) in the active center of human RNA polymerase II. Nucleic Acids Res. 2005;33(8):2629–2639. doi: 10.1093/nar/gki570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer D, Hain J, Thuriaux P, Zillig W. Transcription in archaea: similarity to that in eucarya. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science (Wash DC) 2002a;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science (Wash DC) 2002b;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- Nedialkov YA, Gong XQ, Hovde SL, Yamaguchi Y, Handa H, Geiger JH, Yan H, Burton ZF. NTP-driven translocation by human RNA polymerase II. J Biol Chem. 2003;278:18303–18312. doi: 10.1074/jbc.M301103200. [DOI] [PubMed] [Google Scholar]
- Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- Reines D. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem. 1992;267:3795–3800. [PMC free article] [PubMed] [Google Scholar]
- Rivetti C, Guthold M, Bustamante C. Wrapping of DNA around the E. coli RNA polymerase open promoter complex. EMBO J. 1999;18:4464–4475. doi: 10.1093/emboj/18.16.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivetti C, Codeluppi S, Dieci G, Bustamante C. Visualizing RNA extrusion and DNA wrapping in transcription elongation complexes of bacterial and eukaryotic RNA polymerases. J Mol Biol. 2003;326:1413–1426. doi: 10.1016/s0022-2836(03)00042-1. [DOI] [PubMed] [Google Scholar]
- Robert F, Douziech M, Forget D, Egly JM, Greenblatt J, Burton ZF, Coulombe B. Wrapping of promoter DNA around the RNA polymerase II initiation complex induced by TFIIF. Mol Cell. 1998;2:341–351. doi: 10.1016/s1097-2765(00)80278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu Rev Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- Sluder AE, Price DH, Greenleaf AL. Elongation by Drosophila RNA polymerase II. Transcription of 3′-extended DNA templates. J Biol Chem. 1988;263:9917–9925. [PubMed] [Google Scholar]
- Steitz TA. Structural biology: a mechanism for all polymerases. Nature (London) 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- Sosunova E, Sosunov V, Kozlov M, Nikiforov V, Goldfarb A, Mustaev A. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc Natl Acad Sci USA. 2003;100:15469–15474. doi: 10.1073/pnas.2536698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surratt CK, Milan SC, Chamberlin MJ. Spontaneous cleavage of RNA in ternary complexes of Escherichia coli RNA polymerase and its significance for the mechanism of transcription. Proc Natl Acad Sci USA. 1991;88:7983–7987. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature (London) 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- Vo NV, Hsu LM, Kane CM, Chamberlin MJ. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 3 Influences of individual DNA elements within the promoter recognition region on abortive initiation and promoter escape. Biochemistry. 2003;42:3798–3811. doi: 10.1021/bi026962v. [DOI] [PubMed] [Google Scholar]
- Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science (Wash DC) 2004a;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004b;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Wind M, Reines D. Transcription elongation factor SII. BioEssays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik NA, Young RA. RNA polymerase II: subunit structure and function. Trends Biochem Sci. 1990;15:347–351. doi: 10.1016/0968-0004(90)90074-l. [DOI] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]