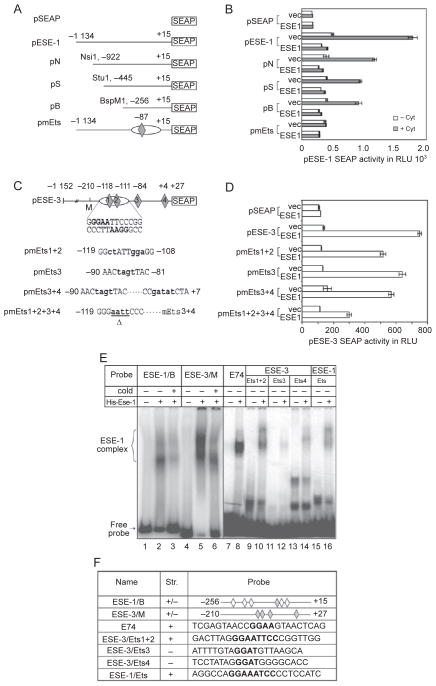
Figure 7.
Deletion and mutation analyses of the ESE-1 and ESE-3 promoters. (A) ESE-1 promoter deletion and mutant reporter plasmids. pSEAP is a vector plasmid containing a promoterless SEAP reporter gene. pESE-1-SEAP is a reporter plasmid containing the full-length promoter. pN, pS and pB are promoter deletion reporter plasmids that are derived from pESE-1-SEAP using restriction enzymes as indicated. pmEts is a full-length promoter reporter with the Ets/NF-κB site mutated. The oval and diamond represent the NF-κB and Ets binding sites, respectively. (B) ESE-1 reporter expression in A549 cells co-transfected with 1 μg of an ESE-1 promoter reporter plasmid and 0.5 μg of an ESE-1 expression plasmid or empty vector. At 24 h after the transfection, the cells were cultured in new media with or without the addition of IL-1β and TNF-α (at 10 ng/ml each) for 24 h. (C) Human ESE-3 promoter wild-type and mutant reporter plasmids. pESE-3-SEAP is a full-length ESE-3 promoter reporter plasmid. The oval and diamonds represent the NF-κB and Ets binding sites, respectively. pmEts1+2, pmEts3, pmEts3+4 and pmEts1+2+3+4 are mutant ESE-3 promoter plasmids with the mutated nucleotides shown in bold. △ indicates deletion of nucleotides. (D) ESE-3 reporter expression in A549 cells co-transfected with 1 μg of an ESE-3 promoter reporter plasmid and 0.5 μg of an ESE-1 expression plasmid or pCDNA3 vector. SEAP activity was measured as relative light units using a luminometer. Values shown are the mean ± SEM (n = 3) from one representative of three independent experiments with similar results. (E) EMSA of ESE-1 protein binding to ESE-1 or ESE-3 promoter fragments or double-stranded oligo nucleotides containing putative Ets binding sites. Full-length ESE-1 fusion protein with a His-tag was expressed and purified fromE. coli. E74 probe was used as a positive control for ESE-1 binding. Bracket indicates specific DNA-protein complexes. ‘Cold’ refers to the addition of a 500-fold unlabelled competitor probe. (F) Probes used in the EMSA. Gray diamonds represent the putative Ets sites that are mutated in (A) and (C), white diamonds represent the putative Ets sites (GGAA/T) that are not characterized, and ‘+’ and ‘−’ represent the top and bottom strands, respectively.