Figure 4. Mesenchymal differentiation elicited by CR-1 overexpression is mediated through parallel actions of AKT and FGFR signals and changes in expression of EMT-TF genes.
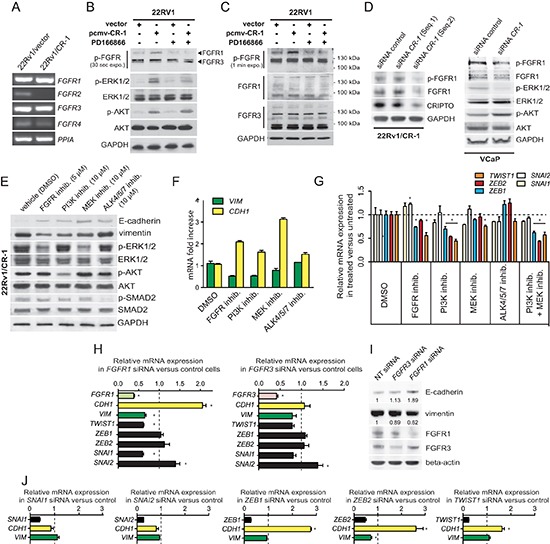
(A) RT-PCR analysis depicting the mRNA levels of the FGFR genes in 22Rv1/vector and 22Rv1/CR-1 cells. The housekeeping gene PPIA is used as an internal control (B) Immunoblots assessing the effect CR-1 cDNA transfection or control vector in 22Rv1, left untreated, or treated with FGFR inhib. (PD166866; 5 μmol/L). Phosphorylation of AKT, ERK1/2, and FGFR was analyzed. (C) The same samples were subjected to immunoblotting for FGFR1 and FGFR3 proteins. (D) Immunoblots made against the indicated proteins from 22Rv1/CR-1 and VCaP cells treated with control or two siRNAs targeting CR-1. (E) 22Rv1/CR-1 cells were treated with inhibitors to PI3K (LY294002, 10 μmol/L), MEK (U0126, 10 μmol/L), FGFR (PD166866, 5 μmol/L), ALK4/5/7 (SB 431542, 10 μmol/L) for 40h and lysates analyzed as above. (F) Parallel qRT-PCR analysis for CDH1 (E-cadherin) and VIM (vimentin) mRNA levels. (G) qRT-PCR revealing perturbations of ZEB1, ZEB2 and TWIST1 levels in inhibitor-treated 22Rv1/CR-1 cells. (H) qRT-PCR analysis for FGFR1, FGFR3, CDH1, VIM, and EMT-TFs following targeting of FGFR1 or FGFR3. (I) Parallel WB analysis for E-cadherin and vimentin. (J) 22Rv1/CR-1 cells were treated with non-targeting siRNAs or siRNAs against ZEB1, ZEB2, SNAI1, SNAI2, or TWIST1, and mRNA levels of CDH1, VIM, and knockdown target genes were assessed by qRT-PCR. Error bars represent n = 3, mean ± sem.
