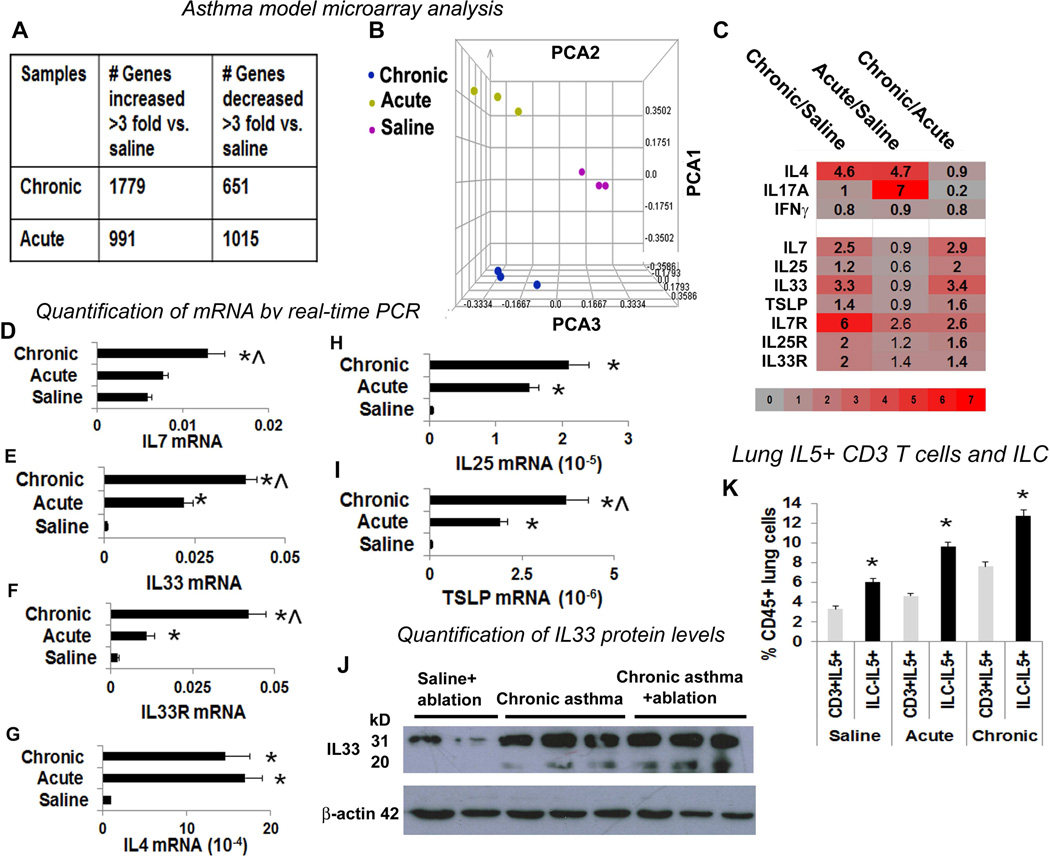
Figure 3. Comparison of the gene expression profile between chronic and acute asthma by microarray.
(A) Microarray analysis of lung tissue obtained from mice with chronic and acute asthma models. N=3 mice/ group. The table shows differentially expressed genes in the asthma models in comparison to saline controls. (B) Principal component analysis (PCA) of the expressed genes demonstrating differences among the asthma models as reflected by their spatial positioning in the 3D space. (C) Heat map of select gene expression changes in chronic and acute asthma as compared to saline controls. (D–I) Measurement of mRNA for select genes by real-time PCR. The mRNA level was normalized to GAPDH mRNA expression. N=5 mice/group, *p<0.05. (J) IL33 levels were confirmed by western blotting to determine the presence of both full length IL33 and the mature cleaved product (N=3). (K) Comparison of lung IL5+CD3 T cells and IL5+ILC2 among the study groups. *P<0.05 as compared to CD3+IL5+ cells in each study group, N=5.