Fig 4. Interference with epigenetically-defined biomarkers perturbs the hESC epigenome.
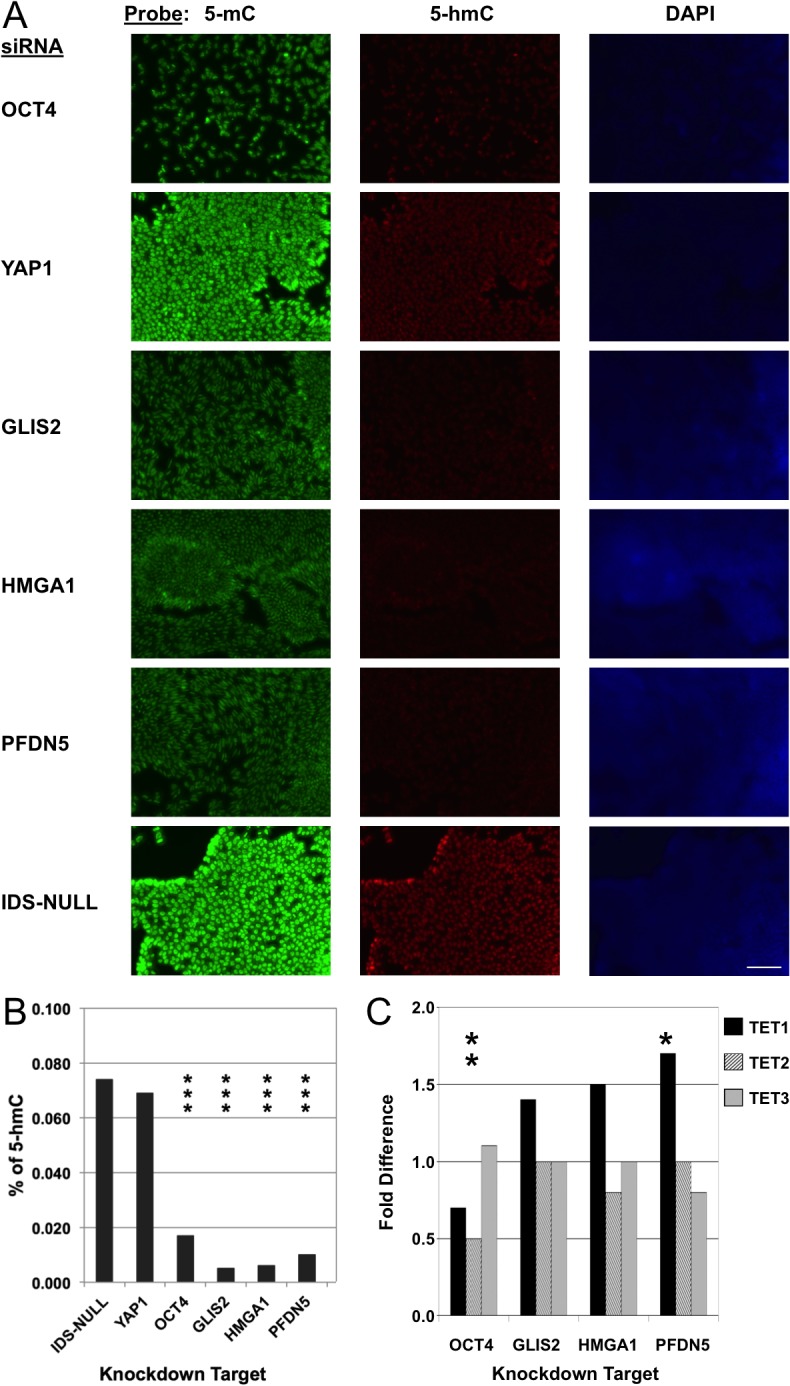
RH1 hESCs were treated with siRNAs as indicated. (A) Immunohistochemical staining for 5-methylcytosine (5-mC) and 5-hydroxymethylcytosine (5-hmC). After knockdown of OCT4, GLIS2, HMGA1 or PFDN5, 5-hmC is difficult to detect. Knockdown of YAP1 or the negative control siRNA (IDS-NULL) had no effect on 5-hmC in hESCs. Scale bar = 100 μm. (B) Quantification of 5-hmC by ELISA as a percentage of total cytosine in gDNA confirming a large (>80%) fall in 5-hmC in hESCs when OCT4, GLIS2, HMGA1 or PFDN5 is knocked down. (C) RT-qPCR data showing fold change in TET1-3 expression on OCT4, GLIS2, HMGA1 or PFDN5 knockdown. TET transcript level changes were mostly not significant or modest (within ~1.5-fold). Asterisks from 1–4 indicate levels of statistical significance, cf. IDS-NULL.
