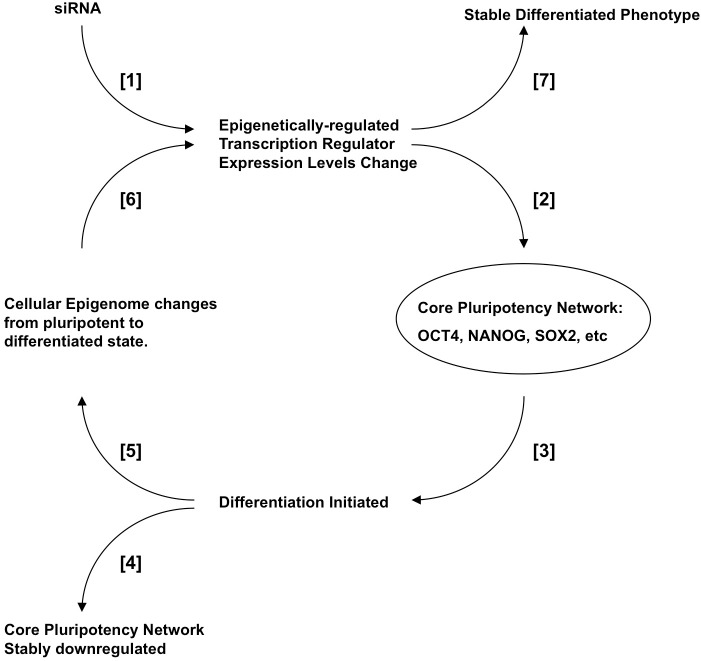
Fig 7. Model of the relationship between epigenetically-regulated hESC biomarkers and the pluripotency transcription system.
Changes in the expression of an epigenetically-regulated transcriptional regulator (e.g. HMGA1, GLIS2, PFDN5) achieved by siRNA transfection [1], or changes in associated CGI methylation [6], feed through the cellular transcription network, resulting in a reduction in core pluripotency transcription factors (OCT4, NANOG, SOX2, [2]) and differentiation [3]. The core pluripotency transcription factors are permanently downregulated [4], and epigenetic changes to gene promoters, CGIs and other regulatory regions of the genome occur to confer stability on the differentiated phenotype and prevent reversion to pluripotency or quasi-pluripotency [5]. These changes confer permanent changes to the expression of the epigenetically regulated transcriptional regulators [6], and thus stabilise the differentiated phenotype [7].