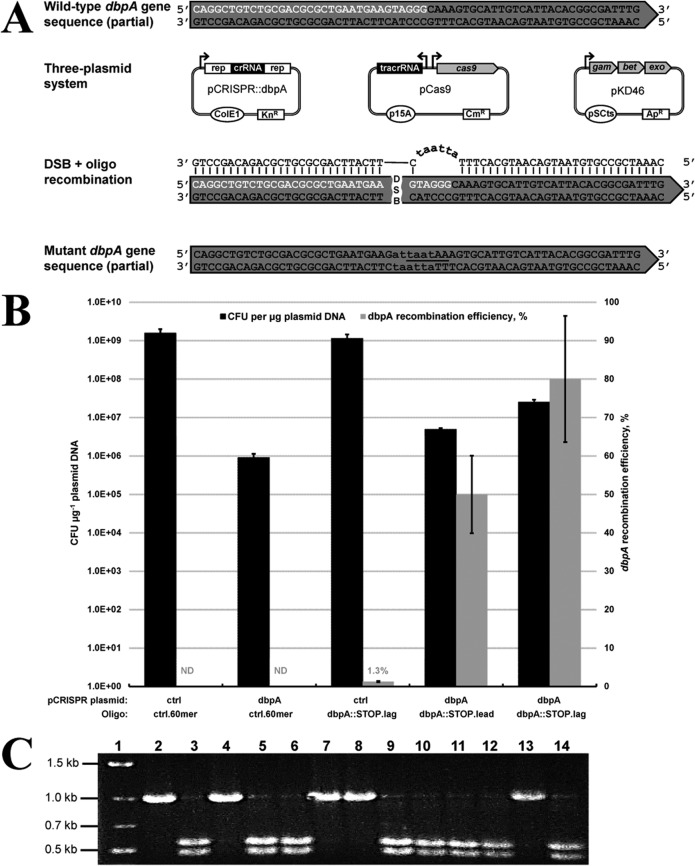
FIG 1.
Single-stranded oligonucleotide recombineering demonstration using the proposed three-plasmid system. (A) Disruption of chromosomal dbpA in E. coli. A 60-nucleotide recombinogenic oligonucleotide targeting the lagging strand of DNA replication (dbpA::STOP.lag) was utilized to disrupt dbpA by introducing six consecutive base pair changes and an AseI recognition sequence (lowercase), generating two consecutive in-frame stop codons (underlined). Sequence corresponding to the dbpA protospacer and PAM are shown in white. Rep corresponds to 36-bp repeats necessary for crRNA processing. (B) Electroporation and dbpA recombination efficiency data resulting from electroporation (pCRISPR::ctrl plus ctrl.60mer), CRISPR/Cas9 (pCRISPR::dbpA plus ctrl.60mer), and recombineering (pCRISPR::ctrl plus dbpA::STOP.lag) controls, as well as CRISPR/Cas9-coupled recombineering using leading or lagging strand oligonucleotides (pCRISPR::dbpA plus dbpA::STOP.lead or dbpA::STOP.lag). Electroporation efficiency is defined as the total number of CFU generated per microgram of plasmid DNA (pCRISPR::ctrl or pCRISPR::dbpA), and recombination efficiency was measured by determining the proportion of mutant colonies following PCR screening and AseI digestion. Recombination efficiency at the dbpA locus was not determined (ND) for electroporation of the nonrecombinogenic control oligonucleotide (ctrl.60mer). Results are averages of at least two independent experiments, and error bars depict standard deviations. (C) Colony PCR and AseI digestion screening of oligonucleotide-mediated dbpA gene disruption. A representative 13 colonies were used as the template in colony PCR with primers dbpA1.Fw and dbpA1.Rv, yielding a product of 1,002 bp. Successful oligonucleotide recombination generates products of 464 bp and 538 bp upon AseI digestion. Lane 1, marker; lanes 2, 4, 7, 8, and 13, negative, unmodified colonies; lanes 3, 5, 6, 9 to 12, and 14, positive, dbpA gene disruption colonies.