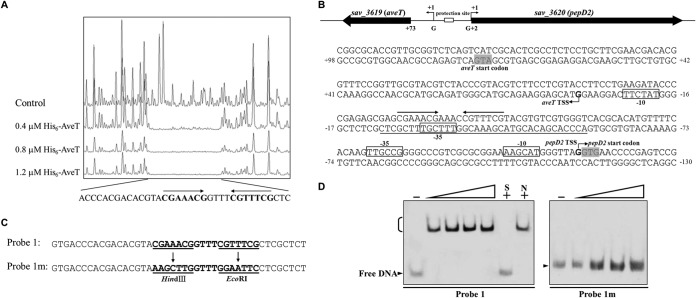
FIG 5.
Determination of AveT-binding site. (A) DNase I footprinting assay of AveT in the aveT-pepD2 intergenic region. Upper fluorogram, control reaction without protein. Protection patterns were obtained with increasing concentrations (0.4, 0.8, 1.2 μM) of the His6-AveT protein. (B) Nucleotide sequences of the aveT-pepD2 promoter region and AveT-binding site. Numbers, distances (in nucleotides) from the TSS of aveT; solid line, AveT-binding site; straight arrows, inverted repeats; bent arrows, TSSs; boxes, putative −10 and −35 regions; shaded areas, translational start codons. (C) Mutations introduced into the 18-bp palindromic sequence. Each probe was 43 bp. Probe 1 was WT DNA containing an intact 18-bp palindromic sequence. Inverted repeats in probe 1 were replaced with HindIII and EcoRI sites to produce mutated probe 1m. Underlining, altered nucleotides. (D) EMSAs using probe 1 and mutated probe 1m. Each reaction mixture contained 0.75 nM labeled probe. The concentrations of the His6-AveT protein for the probes were as follows: for probe 1, 0.1, 0.2, 0.3, and 0.4 μM; for probe 1m, 0.4, 0.8, 1.2, and 1.6 μM.