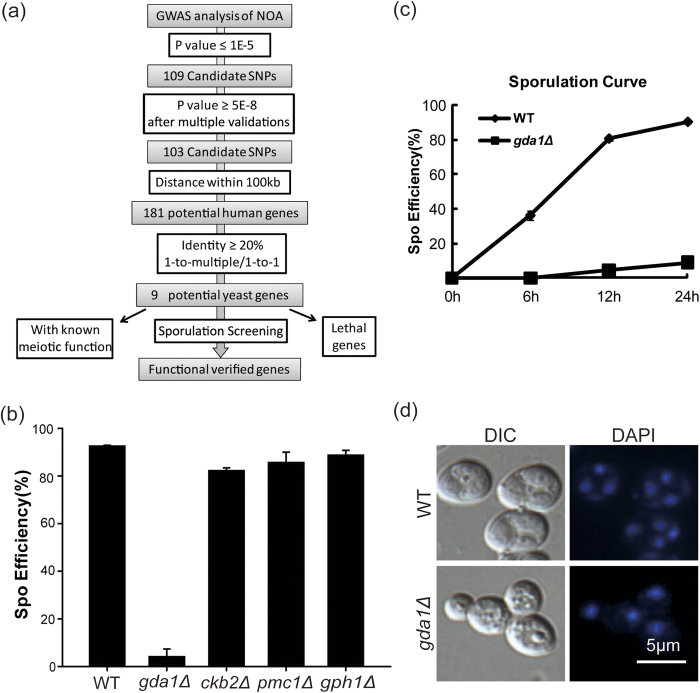
Figure 1. Identification of potential non-obstructive azoospermia pathogenic genes by functional genomic screening in yeast.
(a) Flow chart of the screening strategy. The selection criteria for candidate genes in Saccharomvces cerevisiae included a tSNP with an association P-value < 10−5 and a P-value ≥ 5*10−8 after multiple validations, and human genes flanking the tSNPs within 100 kb, homology type (one to many or one to one) and orthology identity >20% were considered. After eliminating the well-studied genes in meiosis and lethal genes, the candidate genes were screened for their sporulation efficiency after deletion. (b) The sporulation efficiency of the yeast in which candidate genes were deleted. Wild type and candidate gene deletion stains were incubated in sporulation medium for 24 hrs. Sporulation efficiency was the percentage of cells induced to sporulate that became dyads and tetrads by staining with DAPI. (c) The gda1Δ strain showed a decrease in sporulation efficiency compared with the WT strain. A sporulation time course indicated the percentage of cells/asci with dyads and tetrads in the gda1Δ and WT strains. Diploid yeast cells were deprived of nutrients, induced to enter sporulation synchronously, and stained with DAPI at different times post-induction. (d) WT and gda1Δ spores were stained with DAPI to show the decrease of sporulation efficiency.