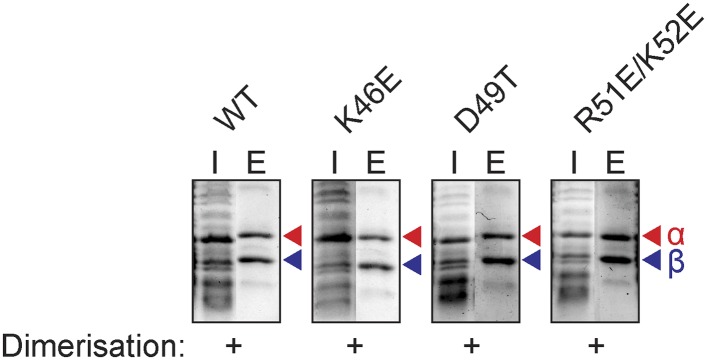
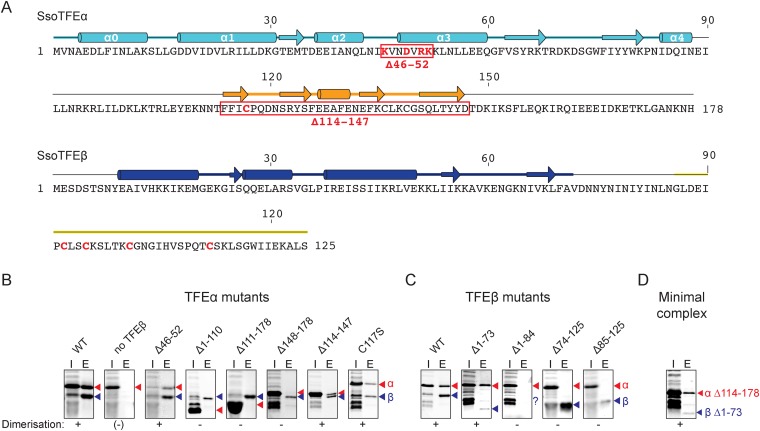
Figure 3. Characterisation of the TFEα/β heterodimerization interface.
(A) Amino acid sequences and secondary structure of TFEα and TFEβ. The α-helices in the TFEα WH domain are numbered according to (Meinhart et al., 2003). Deletion (red boxes) and substitution deletions (highlighted in red) are indicated. (B, C) TFE α/β interaction analysis using co-purification of TFE alpha and his-tagged TFE beta. TFEα (B) and TFEβ-His (C) substitution- and domain deletion variants were co-expressed in E. coli and purified using Nickel-affinity chromatography. Input (heat-stable cell lysate, 25%) (I) and elution fractions (E) were analysed by SDS-PAGE and Coomassie staining. Red and blue triangles indicate the position of the respective mutant variants of TFEα and TFEβ-His, respectively. The question mark denotes that the TFEβ Δ1–84 variant is probably instable. Note that the TFEα expression levels are higher than TFEβ, and that the expression levels of TFEβ mutants are lower than WT, which in some cases make it difficult to discern in the input fractions. (D) Minimal heterodimeric TFEα/β complex consisting of TFEα Δ114–178 and TFEβ Δ1–73.
Figure 3—figure supplement 1. Additional mutants of the N-terminal tip of α-helix 3 of TFEα and their effect on dimerization with TFEβ.