Figure 5. TRPM7 controls progenitor-like features of neuroblastic neuroblastoma cells at the gene expression level.
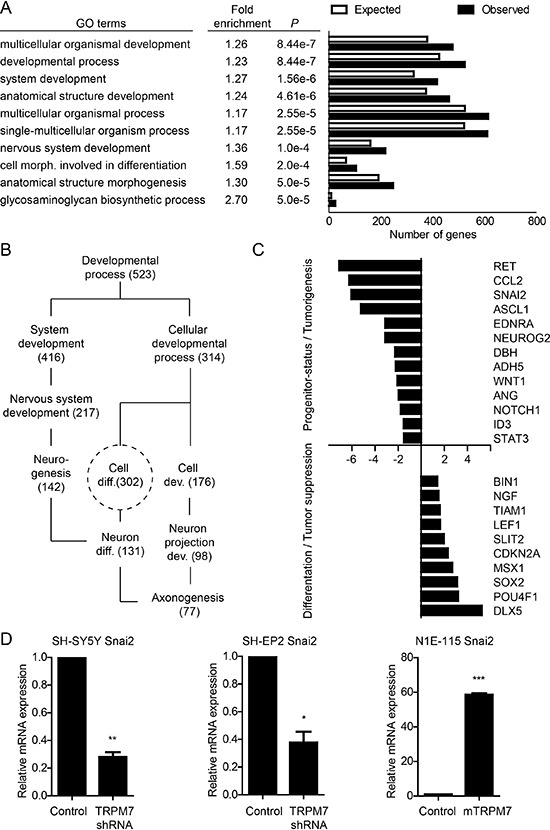
(A) TRPM7 controls developmental gene expression programs. GO-term analysis was performed on genes that were differentially expressed (p < 0.05) in SH-SY5Y TRPM7 shRNA cells, when compared to SH-SY5Y control cells. Presented are the 10 most significantly enriched (P) GO-terms within the subroot ‘Biological Process’. Fold enrichment is the ratio between observed and expected genes within a category. Number of expected genes is based on the number of protein encoding genes within a category. (B) TRPM7 shRNA affects genes that control neuronal differentiation. Simplified representation of GO-term relationships involved in neuronal differentiation, and the number of genes observed per category. (C) TRPM7 shRNA impairs progenitor-status and tumorigenicity of neuroblastic SH-SY5Y neuroblastoma cells. Selection of differentially regulated genes within the ‘Cell Differentiation’ category that are known to control neural crest formation, delamination and differentiation, and neuroblastoma progression (Table 1). X-axis represents fold difference in normalized expression levels between control and TRPM7 shRNA SH-SY5Y cells. (D) TRPM7 drives expression of SNAI2 transcription factor in human and mouse neuroblastoma cell lines. SNAI2 expression levels were determined by quantitative RT-PCR in SH-SY5Y and SH-EP2 control and TRPM7 shRNA cells. Additionally, SNAI2 expression levels were quantified in N1E-115 mouse neuroblastoma control and TRPM7 overexpressing cells. Data represents mean expression levels ± SEM (n = 3) that are normalized to GAPDH housekeeping gene expression. SNAI2 expression in control cells is set to 1. *p < 0.05, **p < 0.01, ***p < 0.001.
