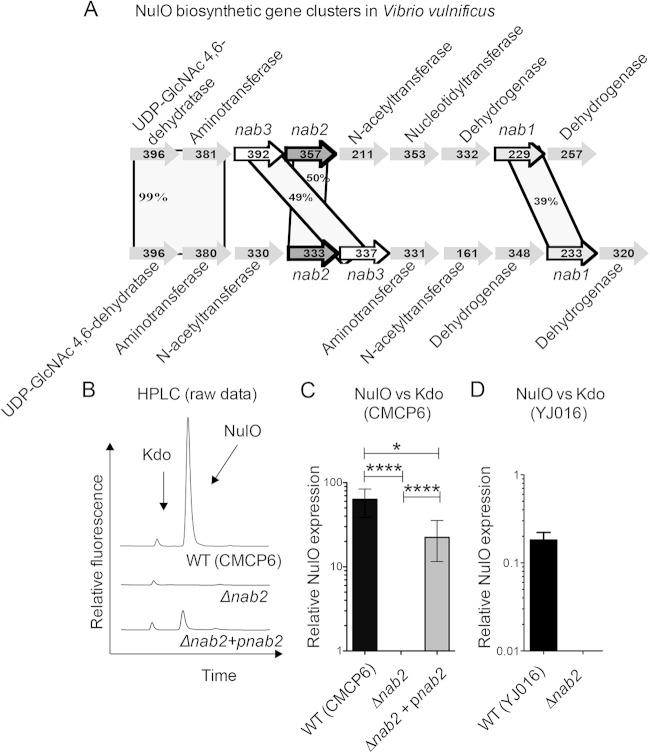
FIG 1.
V. vulnificus nab genetic organization and role of nab2 in NulO biosynthesis. (A) Schematic of the V. vulnificus NulO biosynthetic clusters in YJ016 and CMCP6. Open reading frames and the direction of transcription are designated by arrows. nab1 (encoding a CMP-neuraminic acid synthase homolog), nab2 (encoding an N-acetylneuraminic acid synthase homolog), and nab3 (encoding a UDP-N-acetylglucosamine 2-epimerase homolog) are highlighted. Percentages in shaded areas indicate amino acid identities of translated genes. (B) DMB-HPLC analysis of the CMCP6 WT and isogenic Δnab2 and complemented (Δnab2+pnab2) strains confirmed that nab2 encodes the V. vulnificus NulO synthase. (C and D) NulO expression was normalized to that of 3-deoxy-d-manno-octulosonic acid (Kdo) for quantitative comparisons of NulO expression between strains. Error bars represent standard deviations for three experiments. One-way analysis of variance (ANOVA) with the Bonferroni multiple-comparison posttest (P < 0.05) was used to examine the statistical significance of differences in NulO production. *, P < 0.05; ****, P < 0.0001.