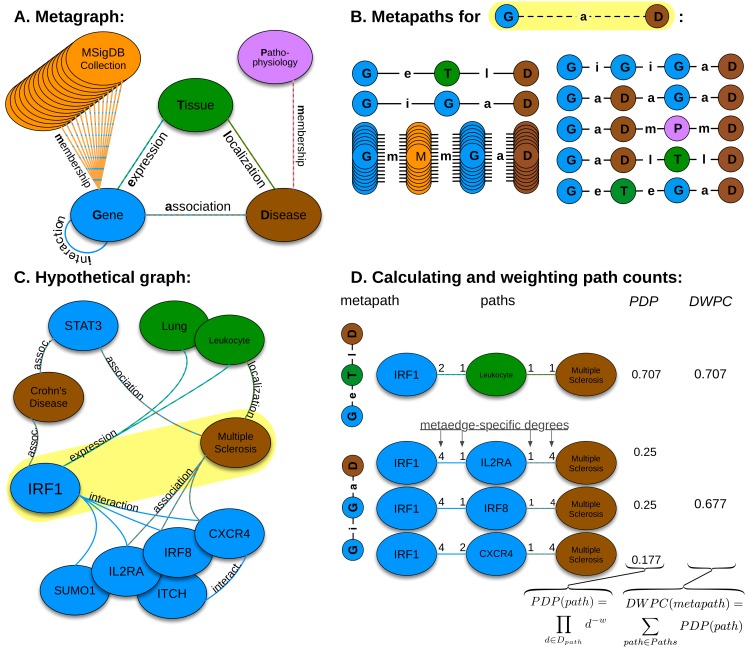
Fig 2. Heterogeneous network edge prediction methodology.
A) We constructed the network according to a schema, called a metagraph, which is composed of metanodes (node types) and metaedges (edge types). B) The network topology connecting a gene and disease node is measured along metapaths (types of paths). Starting on Gene and ending on Disease, all metapaths length three or less are computed by traversing the metagraph. C) A hypothetical graph subset showing select nodes and edges surrounding IRF1 and multiple sclerosis. To characterize this relationship, features are computed that measure the prevalence of a specific metapath between IRF1 and multiple sclerosis. D) Two features (for the GeTlD and GiGaD metapaths) are calculated to describe the relationship between IRF1 and multiple sclerosis. The metric underlying the features is degree-weighted path count (DWPC). First, for the specified metapath, all paths are extracted from the network. Next, each path receives a path-degree product (PDP) measuring its specificity (calculated from node-degrees along the path, D path). This step requires a damping exponent (here w = 0.5), which adjusts how severely high-degree paths are downweighted. Finally, the path-degree products are summed to produce the DWPC.