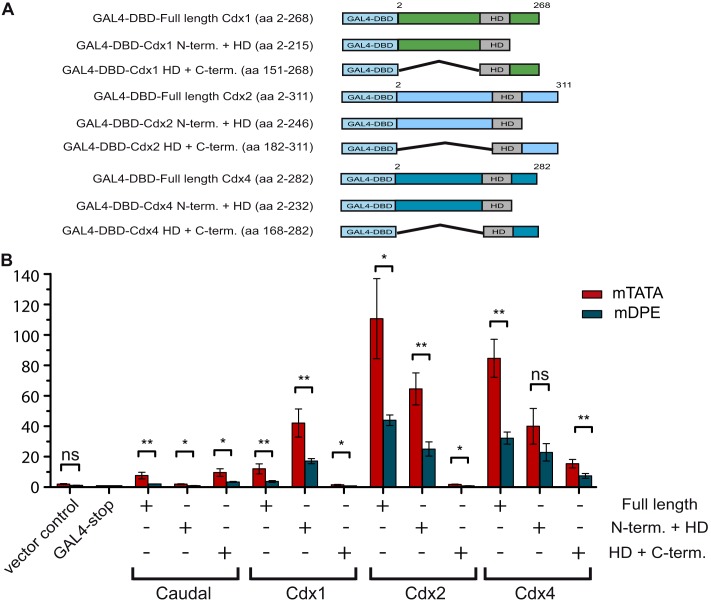
FIGURE 4.
The region containing the homeodomain and the N terminus of mouse Cdx1, Cdx2, and Cdx4 is sufficient to convey core promoter-preferential activation. A, schematic representation of the GAL4 DNA-binding domain fused to various regions of Cdx1, Cdx2, and Cdx4. B, Drosophila S2R+ cells were co-transfected with the indicated GAL4 DNA-binding domain fusion protein expression vector as well as a firefly luciferase reporter gene driven by five GAL4 sites upstream of a ftz promoter containing either an mTATA or mDPE motif. To normalize for variations in transfection efficiency, cells were co-transfected with a Pol III-Renilla luciferase control plasmid. Cell extracts were assayed for Dual-Luciferase activity. The activities are reported relative to the activity of promoters that were co-transfected with the GAL4-stop expression plasmid, which were defined to be 1. The graph represents an average of three independent experiments. Error bars represent S.E. ns, p > 0.05; *, 0.01 ≤ p ≤ 0.05; **, 0.001 ≤ p ≤ 0.01.